Imbalanced classification involves developing predictive models on classification datasets that have a severe class imbalance.
The challenge of working with imbalanced datasets is that most machine learning techniques will ignore, and in turn have poor performance on, the minority class, although typically it is performance on the minority class that is most important.
One approach to addressing imbalanced datasets is to oversample the minority class. The simplest approach involves duplicating examples in the minority class, although these examples don’t add any new information to the model. Instead, new examples can be synthesized from the existing examples. This is a type of data augmentation for the minority class and is referred to as the Synthetic Minority Oversampling Technique, or SMOTE for short.
In this tutorial, you will discover the SMOTE for oversampling imbalanced classification datasets.
After completing this tutorial, you will know:
- How the SMOTE synthesizes new examples for the minority class.
- How to correctly fit and evaluate machine learning models on SMOTE-transformed training datasets.
- How to use extensions of the SMOTE that generate synthetic examples along the class decision boundary.
Kick-start your project with my new book Imbalanced Classification with Python, including step-by-step tutorials and the Python source code files for all examples.
Let’s get started.
- Updated Jan/2021: Updated links for API documentation.

SMOTE Oversampling for Imbalanced Classification with Python
Photo by Victor U, some rights reserved.
Tutorial Overview
This tutorial is divided into five parts; they are:
- Synthetic Minority Oversampling Technique
- Imbalanced-Learn Library
- SMOTE for Balancing Data
- SMOTE for Classification
- SMOTE With Selective Synthetic Sample Generation
- Borderline-SMOTE
- Borderline-SMOTE SVM
- Adaptive Synthetic Sampling (ADASYN)
Synthetic Minority Oversampling Technique
A problem with imbalanced classification is that there are too few examples of the minority class for a model to effectively learn the decision boundary.
One way to solve this problem is to oversample the examples in the minority class. This can be achieved by simply duplicating examples from the minority class in the training dataset prior to fitting a model. This can balance the class distribution but does not provide any additional information to the model.
An improvement on duplicating examples from the minority class is to synthesize new examples from the minority class. This is a type of data augmentation for tabular data and can be very effective.
Perhaps the most widely used approach to synthesizing new examples is called the Synthetic Minority Oversampling TEchnique, or SMOTE for short. This technique was described by Nitesh Chawla, et al. in their 2002 paper named for the technique titled “SMOTE: Synthetic Minority Over-sampling Technique.”
SMOTE works by selecting examples that are close in the feature space, drawing a line between the examples in the feature space and drawing a new sample at a point along that line.
Specifically, a random example from the minority class is first chosen. Then k of the nearest neighbors for that example are found (typically k=5). A randomly selected neighbor is chosen and a synthetic example is created at a randomly selected point between the two examples in feature space.
… SMOTE first selects a minority class instance a at random and finds its k nearest minority class neighbors. The synthetic instance is then created by choosing one of the k nearest neighbors b at random and connecting a and b to form a line segment in the feature space. The synthetic instances are generated as a convex combination of the two chosen instances a and b.
— Page 47, Imbalanced Learning: Foundations, Algorithms, and Applications, 2013.
This procedure can be used to create as many synthetic examples for the minority class as are required. As described in the paper, it suggests first using random undersampling to trim the number of examples in the majority class, then use SMOTE to oversample the minority class to balance the class distribution.
The combination of SMOTE and under-sampling performs better than plain under-sampling.
— SMOTE: Synthetic Minority Over-sampling Technique, 2011.
The approach is effective because new synthetic examples from the minority class are created that are plausible, that is, are relatively close in feature space to existing examples from the minority class.
Our method of synthetic over-sampling works to cause the classifier to build larger decision regions that contain nearby minority class points.
— SMOTE: Synthetic Minority Over-sampling Technique, 2011.
A general downside of the approach is that synthetic examples are created without considering the majority class, possibly resulting in ambiguous examples if there is a strong overlap for the classes.
Now that we are familiar with the technique, let’s look at a worked example for an imbalanced classification problem.
Imbalanced-Learn Library
In these examples, we will use the implementations provided by the imbalanced-learn Python library, which can be installed via pip as follows:
|
1 |
sudo pip install imbalanced-learn |
You can confirm that the installation was successful by printing the version of the installed library:
|
1 2 3 |
# check version number import imblearn print(imblearn.__version__) |
Running the example will print the version number of the installed library; for example:
|
1 |
0.5.0 |
Want to Get Started With Imbalance Classification?
Take my free 7-day email crash course now (with sample code).
Click to sign-up and also get a free PDF Ebook version of the course.
SMOTE for Balancing Data
In this section, we will develop an intuition for the SMOTE by applying it to an imbalanced binary classification problem.
First, we can use the make_classification() scikit-learn function to create a synthetic binary classification dataset with 10,000 examples and a 1:100 class distribution.
|
1 2 3 4 |
... # define dataset X, y = make_classification(n_samples=10000, n_features=2, n_redundant=0, n_clusters_per_class=1, weights=[0.99], flip_y=0, random_state=1) |
We can use the Counter object to summarize the number of examples in each class to confirm the dataset was created correctly.
|
1 2 3 4 |
... # summarize class distribution counter = Counter(y) print(counter) |
Finally, we can create a scatter plot of the dataset and color the examples for each class a different color to clearly see the spatial nature of the class imbalance.
|
1 2 3 4 5 6 7 |
... # scatter plot of examples by class label for label, _ in counter.items(): row_ix = where(y == label)[0] pyplot.scatter(X[row_ix, 0], X[row_ix, 1], label=str(label)) pyplot.legend() pyplot.show() |
Tying this all together, the complete example of generating and plotting a synthetic binary classification problem is listed below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 |
# Generate and plot a synthetic imbalanced classification dataset from collections import Counter from sklearn.datasets import make_classification from matplotlib import pyplot from numpy import where # define dataset X, y = make_classification(n_samples=10000, n_features=2, n_redundant=0, n_clusters_per_class=1, weights=[0.99], flip_y=0, random_state=1) # summarize class distribution counter = Counter(y) print(counter) # scatter plot of examples by class label for label, _ in counter.items(): row_ix = where(y == label)[0] pyplot.scatter(X[row_ix, 0], X[row_ix, 1], label=str(label)) pyplot.legend() pyplot.show() |
Running the example first summarizes the class distribution, confirms the 1:100 ratio, in this case with about 9,900 examples in the majority class and 100 in the minority class.
|
1 |
Counter({0: 9900, 1: 100}) |
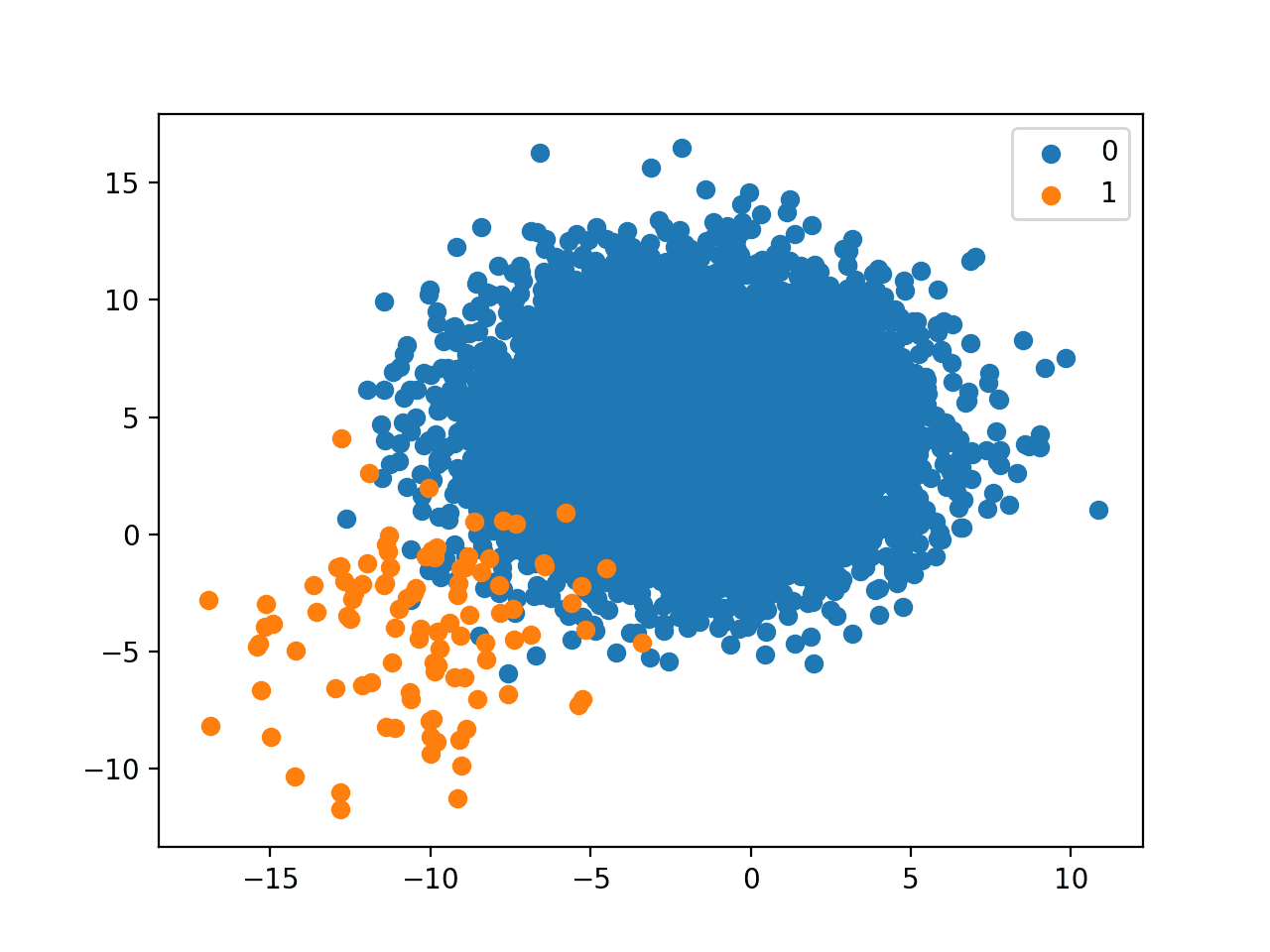
A scatter plot of the dataset is created showing the large mass of points that belong to the majority class (blue) and a small number of points spread out for the minority class (orange). We can see some measure of overlap between the two classes.
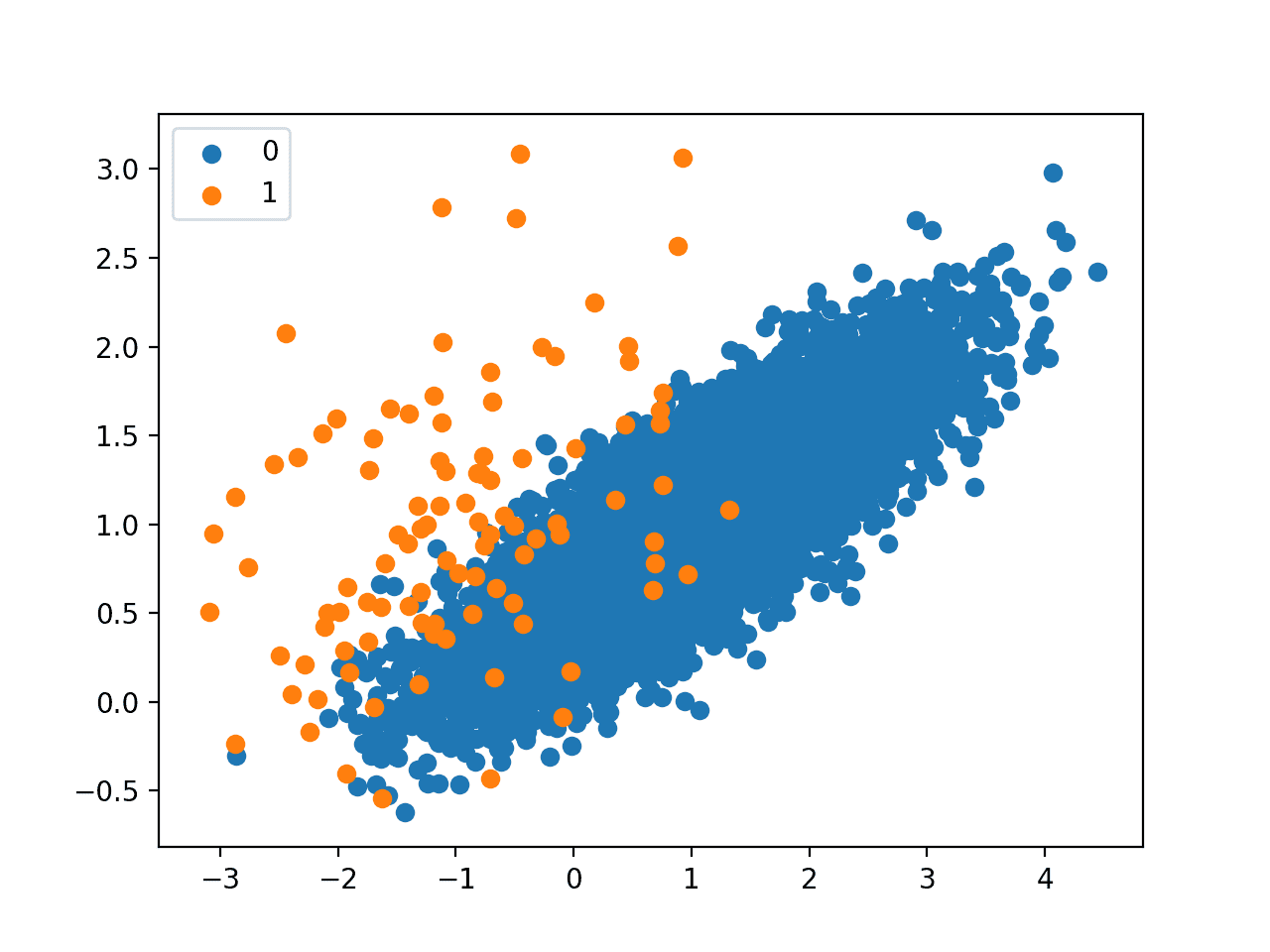
Scatter Plot of Imbalanced Binary Classification Problem
Next, we can oversample the minority class using SMOTE and plot the transformed dataset.
We can use the SMOTE implementation provided by the imbalanced-learn Python library in the SMOTE class.
The SMOTE class acts like a data transform object from scikit-learn in that it must be defined and configured, fit on a dataset, then applied to create a new transformed version of the dataset.
For example, we can define a SMOTE instance with default parameters that will balance the minority class and then fit and apply it in one step to create a transformed version of our dataset.
|
1 2 3 4 |
... # transform the dataset oversample = SMOTE() X, y = oversample.fit_resample(X, y) |
Once transformed, we can summarize the class distribution of the new transformed dataset, which would expect to now be balanced through the creation of many new synthetic examples in the minority class.
|
1 2 3 4 |
... # summarize the new class distribution counter = Counter(y) print(counter) |
A scatter plot of the transformed dataset can also be created and we would expect to see many more examples for the minority class on lines between the original examples in the minority class.
Tying this together, the complete examples of applying SMOTE to the synthetic dataset and then summarizing and plotting the transformed result is listed below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 |
# Oversample and plot imbalanced dataset with SMOTE from collections import Counter from sklearn.datasets import make_classification from imblearn.over_sampling import SMOTE from matplotlib import pyplot from numpy import where # define dataset X, y = make_classification(n_samples=10000, n_features=2, n_redundant=0, n_clusters_per_class=1, weights=[0.99], flip_y=0, random_state=1) # summarize class distribution counter = Counter(y) print(counter) # transform the dataset oversample = SMOTE() X, y = oversample.fit_resample(X, y) # summarize the new class distribution counter = Counter(y) print(counter) # scatter plot of examples by class label for label, _ in counter.items(): row_ix = where(y == label)[0] pyplot.scatter(X[row_ix, 0], X[row_ix, 1], label=str(label)) pyplot.legend() pyplot.show() |
Running the example first creates the dataset and summarizes the class distribution, showing the 1:100 ratio.
Then the dataset is transformed using the SMOTE and the new class distribution is summarized, showing a balanced distribution now with 9,900 examples in the minority class.
|
1 2 |
Counter({0: 9900, 1: 100}) Counter({0: 9900, 1: 9900}) |
Finally, a scatter plot of the transformed dataset is created.
It shows many more examples in the minority class created along the lines between the original examples in the minority class.
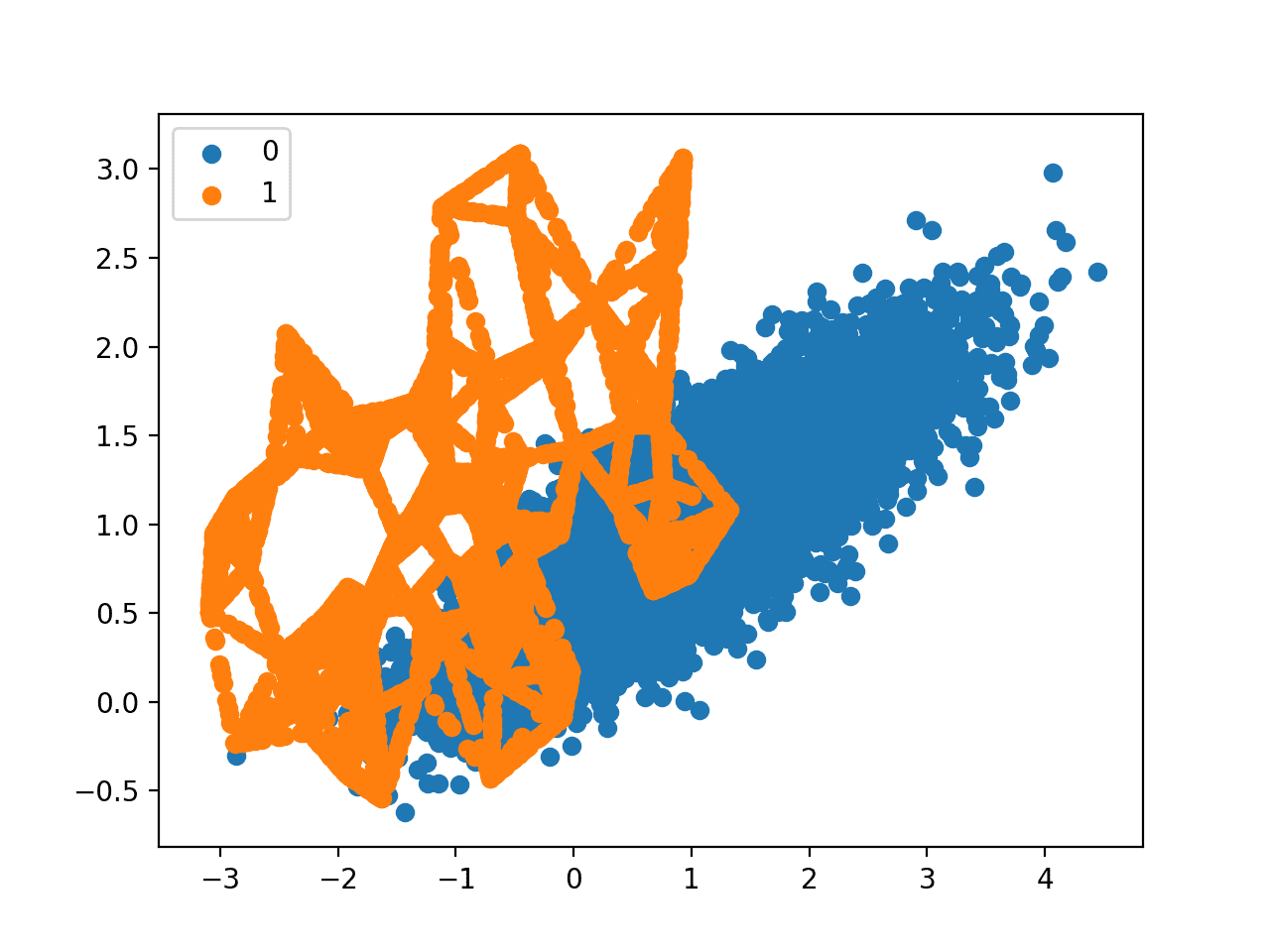
Scatter Plot of Imbalanced Binary Classification Problem Transformed by SMOTE
The original paper on SMOTE suggested combining SMOTE with random undersampling of the majority class.
The imbalanced-learn library supports random undersampling via the RandomUnderSampler class.
We can update the example to first oversample the minority class to have 10 percent the number of examples of the majority class (e.g. about 1,000), then use random undersampling to reduce the number of examples in the majority class to have 50 percent more than the minority class (e.g. about 2,000).
To implement this, we can specify the desired ratios as arguments to the SMOTE and RandomUnderSampler classes; for example:
|
1 2 3 |
... over = SMOTE(sampling_strategy=0.1) under = RandomUnderSampler(sampling_strategy=0.5) |
We can then chain these two transforms together into a Pipeline.
The Pipeline can then be applied to a dataset, performing each transformation in turn and returning a final dataset with the accumulation of the transform applied to it, in this case oversampling followed by undersampling.
|
1 2 3 |
... steps = [('o', over), ('u', under)] pipeline = Pipeline(steps=steps) |
The pipeline can then be fit and applied to our dataset just like a single transform:
|
1 2 3 |
... # transform the dataset X, y = pipeline.fit_resample(X, y) |
We can then summarize and plot the resulting dataset.
We would expect some SMOTE oversampling of the minority class, although not as much as before where the dataset was balanced. We also expect fewer examples in the majority class via random undersampling.
Tying this all together, the complete example is listed below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 |
# Oversample with SMOTE and random undersample for imbalanced dataset from collections import Counter from sklearn.datasets import make_classification from imblearn.over_sampling import SMOTE from imblearn.under_sampling import RandomUnderSampler from imblearn.pipeline import Pipeline from matplotlib import pyplot from numpy import where # define dataset X, y = make_classification(n_samples=10000, n_features=2, n_redundant=0, n_clusters_per_class=1, weights=[0.99], flip_y=0, random_state=1) # summarize class distribution counter = Counter(y) print(counter) # define pipeline over = SMOTE(sampling_strategy=0.1) under = RandomUnderSampler(sampling_strategy=0.5) steps = [('o', over), ('u', under)] pipeline = Pipeline(steps=steps) # transform the dataset X, y = pipeline.fit_resample(X, y) # summarize the new class distribution counter = Counter(y) print(counter) # scatter plot of examples by class label for label, _ in counter.items(): row_ix = where(y == label)[0] pyplot.scatter(X[row_ix, 0], X[row_ix, 1], label=str(label)) pyplot.legend() pyplot.show() |
Running the example first creates the dataset and summarizes the class distribution.
Next, the dataset is transformed, first by oversampling the minority class, then undersampling the majority class. The final class distribution after this sequence of transforms matches our expectations with a 1:2 ratio or about 2,000 examples in the majority class and about 1,000 examples in the minority class.
|
1 2 |
Counter({0: 9900, 1: 100}) Counter({0: 1980, 1: 990}) |
Finally, a scatter plot of the transformed dataset is created, showing the oversampled minority class and the undersampled majority class.
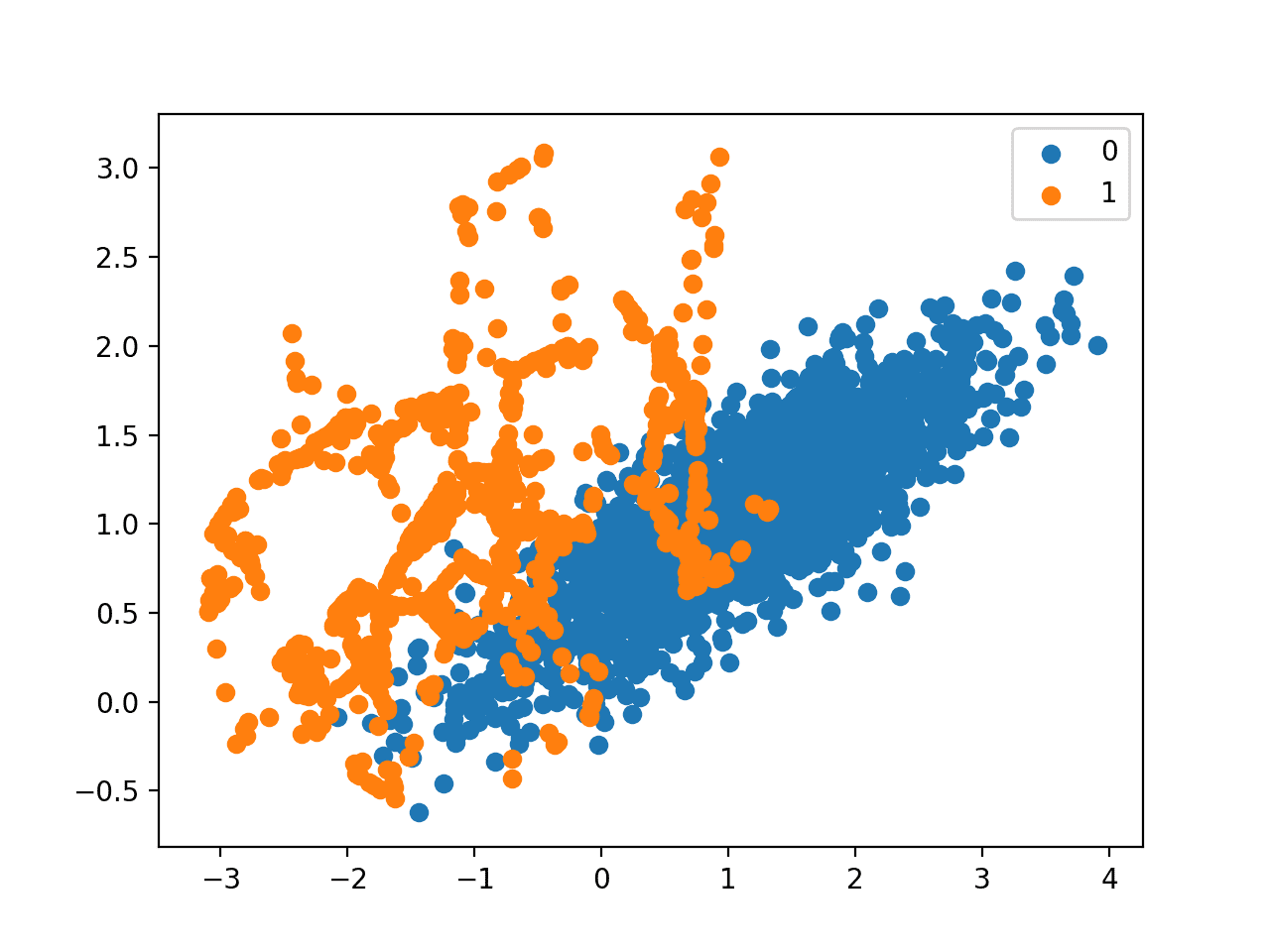
Scatter Plot of Imbalanced Dataset Transformed by SMOTE and Random Undersampling
Now that we are familiar with transforming imbalanced datasets, let’s look at using SMOTE when fitting and evaluating classification models.
SMOTE for Classification
In this section, we will look at how we can use SMOTE as a data preparation method when fitting and evaluating machine learning algorithms in scikit-learn.
First, we use our binary classification dataset from the previous section then fit and evaluate a decision tree algorithm.
The algorithm is defined with any required hyperparameters (we will use the defaults), then we will use repeated stratified k-fold cross-validation to evaluate the model. We will use three repeats of 10-fold cross-validation, meaning that 10-fold cross-validation is applied three times fitting and evaluating 30 models on the dataset.
The dataset is stratified, meaning that each fold of the cross-validation split will have the same class distribution as the original dataset, in this case, a 1:100 ratio. We will evaluate the model using the ROC area under curve (AUC) metric. This can be optimistic for severely imbalanced datasets but will still show a relative change with better performing models.
|
1 2 3 4 5 6 |
... # define model model = DecisionTreeClassifier() # evaluate pipeline cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1) scores = cross_val_score(model, X, y, scoring='roc_auc', cv=cv, n_jobs=-1) |
Once fit, we can calculate and report the mean of the scores across the folds and repeats.
|
1 2 |
... print('Mean ROC AUC: %.3f' % mean(scores)) |
We would not expect a decision tree fit on the raw imbalanced dataset to perform very well.
Tying this together, the complete example is listed below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 |
# decision tree evaluated on imbalanced dataset from numpy import mean from sklearn.datasets import make_classification from sklearn.model_selection import cross_val_score from sklearn.model_selection import RepeatedStratifiedKFold from sklearn.tree import DecisionTreeClassifier # define dataset X, y = make_classification(n_samples=10000, n_features=2, n_redundant=0, n_clusters_per_class=1, weights=[0.99], flip_y=0, random_state=1) # define model model = DecisionTreeClassifier() # evaluate pipeline cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1) scores = cross_val_score(model, X, y, scoring='roc_auc', cv=cv, n_jobs=-1) print('Mean ROC AUC: %.3f' % mean(scores)) |
Running the example evaluates the model and reports the mean ROC AUC.
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
In this case, we can see that a ROC AUC of about 0.76 is reported.
|
1 |
Mean ROC AUC: 0.761 |
Now, we can try the same model and the same evaluation method, although use a SMOTE transformed version of the dataset.
The correct application of oversampling during k-fold cross-validation is to apply the method to the training dataset only, then evaluate the model on the stratified but non-transformed test set.
This can be achieved by defining a Pipeline that first transforms the training dataset with SMOTE then fits the model.
|
1 2 3 4 |
... # define pipeline steps = [('over', SMOTE()), ('model', DecisionTreeClassifier())] pipeline = Pipeline(steps=steps) |
This pipeline can then be evaluated using repeated k-fold cross-validation.
Tying this together, the complete example of evaluating a decision tree with SMOTE oversampling on the training dataset is listed below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 |
# decision tree evaluated on imbalanced dataset with SMOTE oversampling from numpy import mean from sklearn.datasets import make_classification from sklearn.model_selection import cross_val_score from sklearn.model_selection import RepeatedStratifiedKFold from sklearn.tree import DecisionTreeClassifier from imblearn.pipeline import Pipeline from imblearn.over_sampling import SMOTE # define dataset X, y = make_classification(n_samples=10000, n_features=2, n_redundant=0, n_clusters_per_class=1, weights=[0.99], flip_y=0, random_state=1) # define pipeline steps = [('over', SMOTE()), ('model', DecisionTreeClassifier())] pipeline = Pipeline(steps=steps) # evaluate pipeline cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1) scores = cross_val_score(pipeline, X, y, scoring='roc_auc', cv=cv, n_jobs=-1) print('Mean ROC AUC: %.3f' % mean(scores)) |
Running the example evaluates the model and reports the mean ROC AUC score across the multiple folds and repeats.
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
In this case, we can see a modest improvement in performance from a ROC AUC of about 0.76 to about 0.80.
|
1 |
Mean ROC AUC: 0.809 |
As mentioned in the paper, it is believed that SMOTE performs better when combined with undersampling of the majority class, such as random undersampling.
We can achieve this by simply adding a RandomUnderSampler step to the Pipeline.
As in the previous section, we will first oversample the minority class with SMOTE to about a 1:10 ratio, then undersample the majority class to achieve about a 1:2 ratio.
|
1 2 3 4 5 6 7 |
... # define pipeline model = DecisionTreeClassifier() over = SMOTE(sampling_strategy=0.1) under = RandomUnderSampler(sampling_strategy=0.5) steps = [('over', over), ('under', under), ('model', model)] pipeline = Pipeline(steps=steps) |
Tying this together, the complete example is listed below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 |
# decision tree on imbalanced dataset with SMOTE oversampling and random undersampling from numpy import mean from sklearn.datasets import make_classification from sklearn.model_selection import cross_val_score from sklearn.model_selection import RepeatedStratifiedKFold from sklearn.tree import DecisionTreeClassifier from imblearn.pipeline import Pipeline from imblearn.over_sampling import SMOTE from imblearn.under_sampling import RandomUnderSampler # define dataset X, y = make_classification(n_samples=10000, n_features=2, n_redundant=0, n_clusters_per_class=1, weights=[0.99], flip_y=0, random_state=1) # define pipeline model = DecisionTreeClassifier() over = SMOTE(sampling_strategy=0.1) under = RandomUnderSampler(sampling_strategy=0.5) steps = [('over', over), ('under', under), ('model', model)] pipeline = Pipeline(steps=steps) # evaluate pipeline cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1) scores = cross_val_score(pipeline, X, y, scoring='roc_auc', cv=cv, n_jobs=-1) print('Mean ROC AUC: %.3f' % mean(scores)) |
Running the example evaluates the model with the pipeline of SMOTE oversampling and random undersampling on the training dataset.
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
In this case, we can see that the reported ROC AUC shows an additional lift to about 0.83.
|
1 |
Mean ROC AUC: 0.834 |
You could explore testing different ratios of the minority class and majority class (e.g. changing the sampling_strategy argument) to see if a further lift in performance is possible.
Another area to explore would be to test different values of the k-nearest neighbors selected in the SMOTE procedure when each new synthetic example is created. The default is k=5, although larger or smaller values will influence the types of examples created, and in turn, may impact the performance of the model.
For example, we could grid search a range of values of k, such as values from 1 to 7, and evaluate the pipeline for each value.
|
1 2 3 4 5 6 |
... # values to evaluate k_values = [1, 2, 3, 4, 5, 6, 7] for k in k_values: # define pipeline ... |
The complete example is listed below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 |
# grid search k value for SMOTE oversampling for imbalanced classification from numpy import mean from sklearn.datasets import make_classification from sklearn.model_selection import cross_val_score from sklearn.model_selection import RepeatedStratifiedKFold from sklearn.tree import DecisionTreeClassifier from imblearn.pipeline import Pipeline from imblearn.over_sampling import SMOTE from imblearn.under_sampling import RandomUnderSampler # define dataset X, y = make_classification(n_samples=10000, n_features=2, n_redundant=0, n_clusters_per_class=1, weights=[0.99], flip_y=0, random_state=1) # values to evaluate k_values = [1, 2, 3, 4, 5, 6, 7] for k in k_values: # define pipeline model = DecisionTreeClassifier() over = SMOTE(sampling_strategy=0.1, k_neighbors=k) under = RandomUnderSampler(sampling_strategy=0.5) steps = [('over', over), ('under', under), ('model', model)] pipeline = Pipeline(steps=steps) # evaluate pipeline cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1) scores = cross_val_score(pipeline, X, y, scoring='roc_auc', cv=cv, n_jobs=-1) score = mean(scores) print('> k=%d, Mean ROC AUC: %.3f' % (k, score)) |
Running the example will perform SMOTE oversampling with different k values for the KNN used in the procedure, followed by random undersampling and fitting a decision tree on the resulting training dataset.
The mean ROC AUC is reported for each configuration.
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
In this case, the results suggest that a k=3 might be good with a ROC AUC of about 0.84, and k=7 might also be good with a ROC AUC of about 0.85.
This highlights that both the amount of oversampling and undersampling performed (sampling_strategy argument) and the number of examples selected from which a partner is chosen to create a synthetic example (k_neighbors) may be important parameters to select and tune for your dataset.
|
1 2 3 4 5 6 7 |
> k=1, Mean ROC AUC: 0.827 > k=2, Mean ROC AUC: 0.823 > k=3, Mean ROC AUC: 0.834 > k=4, Mean ROC AUC: 0.840 > k=5, Mean ROC AUC: 0.839 > k=6, Mean ROC AUC: 0.839 > k=7, Mean ROC AUC: 0.853 |
Now that we are familiar with how to use SMOTE when fitting and evaluating classification models, let’s look at some extensions of the SMOTE procedure.
SMOTE With Selective Synthetic Sample Generation
We can be selective about the examples in the minority class that are oversampled using SMOTE.
In this section, we will review some extensions to SMOTE that are more selective regarding the examples from the minority class that provide the basis for generating new synthetic examples.
Borderline-SMOTE
A popular extension to SMOTE involves selecting those instances of the minority class that are misclassified, such as with a k-nearest neighbor classification model.
We can then oversample just those difficult instances, providing more resolution only where it may be required.
The examples on the borderline and the ones nearby […] are more apt to be misclassified than the ones far from the borderline, and thus more important for classification.
— Borderline-SMOTE: A New Over-Sampling Method in Imbalanced Data Sets Learning, 2005.
These examples that are misclassified are likely ambiguous and in a region of the edge or border of decision boundary where class membership may overlap. As such, this modified to SMOTE is called Borderline-SMOTE and was proposed by Hui Han, et al. in their 2005 paper titled “Borderline-SMOTE: A New Over-Sampling Method in Imbalanced Data Sets Learning.”
The authors also describe a version of the method that also oversampled the majority class for those examples that cause a misclassification of borderline instances in the minority class. This is referred to as Borderline-SMOTE1, whereas the oversampling of just the borderline cases in minority class is referred to as Borderline-SMOTE2.
Borderline-SMOTE2 not only generates synthetic examples from each example in DANGER and its positive nearest neighbors in P, but also does that from its nearest negative neighbor in N.
— Borderline-SMOTE: A New Over-Sampling Method in Imbalanced Data Sets Learning, 2005.
We can implement Borderline-SMOTE1 using the BorderlineSMOTE class from imbalanced-learn.
We can demonstrate the technique on the synthetic binary classification problem used in the previous sections.
Instead of generating new synthetic examples for the minority class blindly, we would expect the Borderline-SMOTE method to only create synthetic examples along the decision boundary between the two classes.
The complete example of using Borderline-SMOTE to oversample binary classification datasets is listed below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 |
# borderline-SMOTE for imbalanced dataset from collections import Counter from sklearn.datasets import make_classification from imblearn.over_sampling import BorderlineSMOTE from matplotlib import pyplot from numpy import where # define dataset X, y = make_classification(n_samples=10000, n_features=2, n_redundant=0, n_clusters_per_class=1, weights=[0.99], flip_y=0, random_state=1) # summarize class distribution counter = Counter(y) print(counter) # transform the dataset oversample = BorderlineSMOTE() X, y = oversample.fit_resample(X, y) # summarize the new class distribution counter = Counter(y) print(counter) # scatter plot of examples by class label for label, _ in counter.items(): row_ix = where(y == label)[0] pyplot.scatter(X[row_ix, 0], X[row_ix, 1], label=str(label)) pyplot.legend() pyplot.show() |
Running the example first creates the dataset and summarizes the initial class distribution, showing a 1:100 relationship.
The Borderline-SMOTE is applied to balance the class distribution, which is confirmed with the printed class summary.
|
1 2 |
Counter({0: 9900, 1: 100}) Counter({0: 9900, 1: 9900}) |
Finally, a scatter plot of the transformed dataset is created. The plot clearly shows the effect of the selective approach to oversampling. Examples along the decision boundary of the minority class are oversampled intently (orange).
The plot shows that those examples far from the decision boundary are not oversampled. This includes both examples that are easier to classify (those orange points toward the top left of the plot) and those that are overwhelmingly difficult to classify given the strong class overlap (those orange points toward the bottom right of the plot).
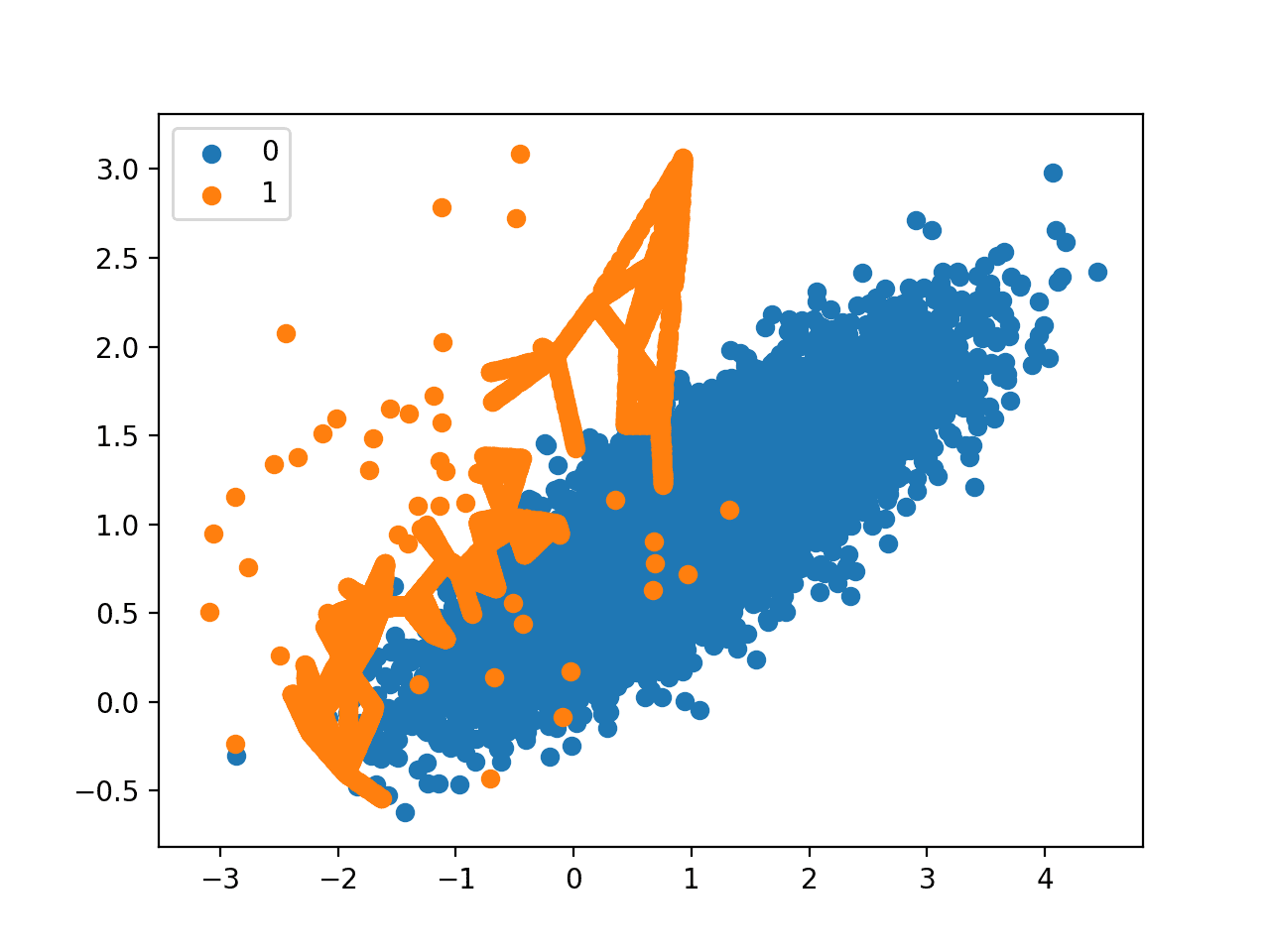
Scatter Plot of Imbalanced Dataset With Borderline-SMOTE Oversampling
Borderline-SMOTE SVM
Hien Nguyen, et al. suggest using an alternative of Borderline-SMOTE where an SVM algorithm is used instead of a KNN to identify misclassified examples on the decision boundary.
Their approach is summarized in the 2009 paper titled “Borderline Over-sampling For Imbalanced Data Classification.” An SVM is used to locate the decision boundary defined by the support vectors and examples in the minority class that close to the support vectors become the focus for generating synthetic examples.
… the borderline area is approximated by the support vectors obtained after training a standard SVMs classifier on the original training set. New instances will be randomly created along the lines joining each minority class support vector with a number of its nearest neighbors using the interpolation
— Borderline Over-sampling For Imbalanced Data Classification, 2009.
In addition to using an SVM, the technique attempts to select regions where there are fewer examples of the minority class and tries to extrapolate towards the class boundary.
If majority class instances count for less than a half of its nearest neighbors, new instances will be created with extrapolation to expand minority class area toward the majority class.
— Borderline Over-sampling For Imbalanced Data Classification, 2009.
This variation can be implemented via the SVMSMOTE class from the imbalanced-learn library.
The example below demonstrates this alternative approach to Borderline SMOTE on the same imbalanced dataset.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 |
# borderline-SMOTE with SVM for imbalanced dataset from collections import Counter from sklearn.datasets import make_classification from imblearn.over_sampling import SVMSMOTE from matplotlib import pyplot from numpy import where # define dataset X, y = make_classification(n_samples=10000, n_features=2, n_redundant=0, n_clusters_per_class=1, weights=[0.99], flip_y=0, random_state=1) # summarize class distribution counter = Counter(y) print(counter) # transform the dataset oversample = SVMSMOTE() X, y = oversample.fit_resample(X, y) # summarize the new class distribution counter = Counter(y) print(counter) # scatter plot of examples by class label for label, _ in counter.items(): row_ix = where(y == label)[0] pyplot.scatter(X[row_ix, 0], X[row_ix, 1], label=str(label)) pyplot.legend() pyplot.show() |
Running the example first summarizes the raw class distribution, then the balanced class distribution after applying Borderline-SMOTE with an SVM model.
|
1 2 |
Counter({0: 9900, 1: 100}) Counter({0: 9900, 1: 9900}) |
A scatter plot of the dataset is created showing the directed oversampling along the decision boundary with the majority class.
We can also see that unlike Borderline-SMOTE, more examples are synthesized away from the region of class overlap, such as toward the top left of the plot.
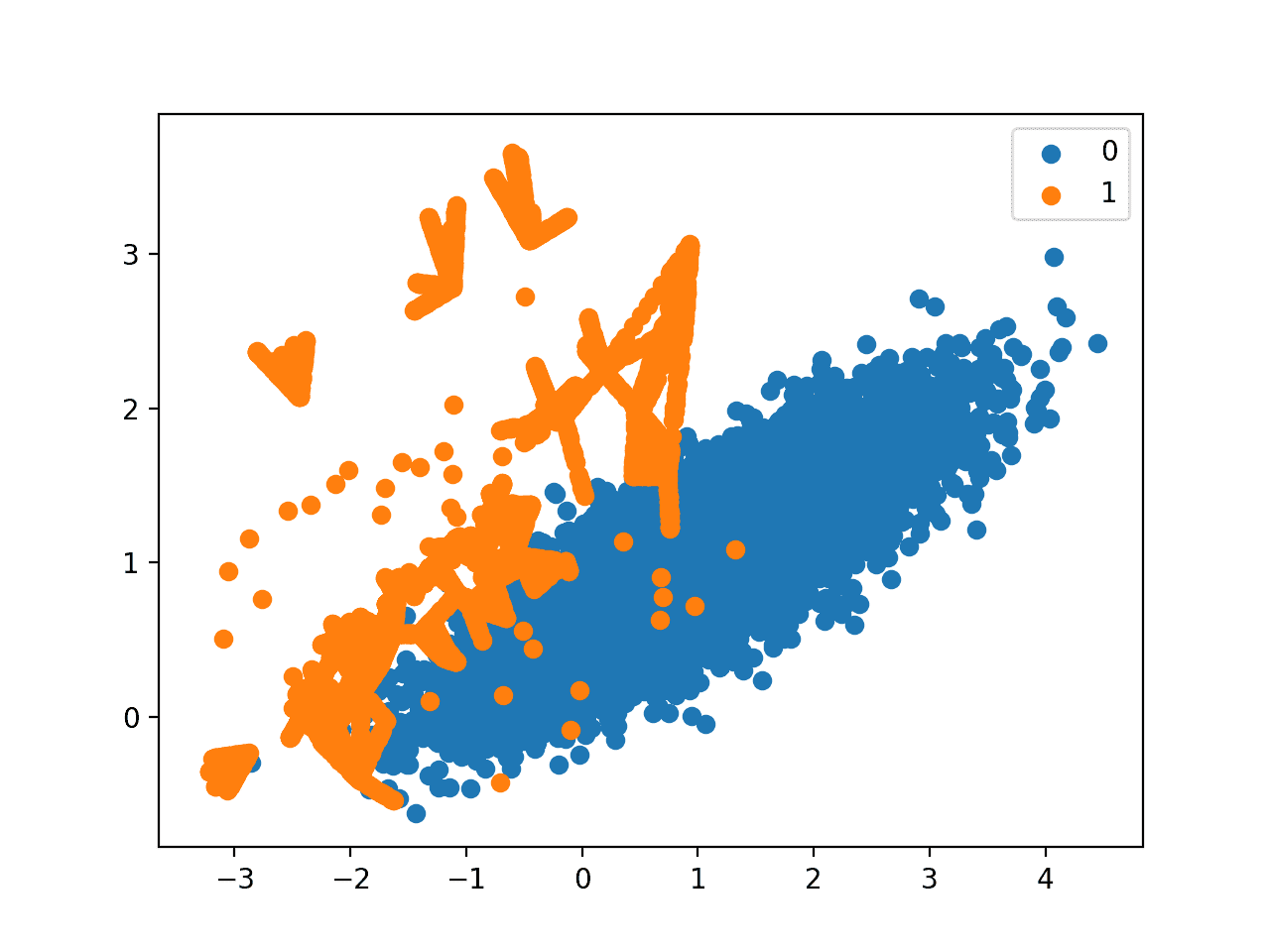
Scatter Plot of Imbalanced Dataset With Borderline-SMOTE Oversampling With SVM
Adaptive Synthetic Sampling (ADASYN)
Another approach involves generating synthetic samples inversely proportional to the density of the examples in the minority class.
That is, generate more synthetic examples in regions of the feature space where the density of minority examples is low, and fewer or none where the density is high.
This modification to SMOTE is referred to as the Adaptive Synthetic Sampling Method, or ADASYN, and was proposed to Haibo He, et al. in their 2008 paper named for the method titled “ADASYN: Adaptive Synthetic Sampling Approach For Imbalanced Learning.”
ADASYN is based on the idea of adaptively generating minority data samples according to their distributions: more synthetic data is generated for minority class samples that are harder to learn compared to those minority samples that are easier to learn.
— ADASYN: Adaptive synthetic sampling approach for imbalanced learning, 2008.
With online Borderline-SMOTE, a discriminative model is not created. Instead, examples in the minority class are weighted according to their density, then those examples with the lowest density are the focus for the SMOTE synthetic example generation process.
The key idea of ADASYN algorithm is to use a density distribution as a criterion to automatically decide the number of synthetic samples that need to be generated for each minority data example.
— ADASYN: Adaptive synthetic sampling approach for imbalanced learning, 2008.
We can implement this procedure using the ADASYN class in the imbalanced-learn library.
The example below demonstrates this alternative approach to oversampling on the imbalanced binary classification dataset.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 |
# Oversample and plot imbalanced dataset with ADASYN from collections import Counter from sklearn.datasets import make_classification from imblearn.over_sampling import ADASYN from matplotlib import pyplot from numpy import where # define dataset X, y = make_classification(n_samples=10000, n_features=2, n_redundant=0, n_clusters_per_class=1, weights=[0.99], flip_y=0, random_state=1) # summarize class distribution counter = Counter(y) print(counter) # transform the dataset oversample = ADASYN() X, y = oversample.fit_resample(X, y) # summarize the new class distribution counter = Counter(y) print(counter) # scatter plot of examples by class label for label, _ in counter.items(): row_ix = where(y == label)[0] pyplot.scatter(X[row_ix, 0], X[row_ix, 1], label=str(label)) pyplot.legend() pyplot.show() |
Running the example first creates the dataset and summarizes the initial class distribution, then the updated class distribution after oversampling was performed.
|
1 2 |
Counter({0: 9900, 1: 100}) Counter({0: 9900, 1: 9899}) |
A scatter plot of the transformed dataset is created. Like Borderline-SMOTE, we can see that synthetic sample generation is focused around the decision boundary as this region has the lowest density.
Unlike Borderline-SMOTE, we can see that the examples that have the most class overlap have the most focus. On problems where these low density examples might be outliers, the ADASYN approach may put too much attention on these areas of the feature space, which may result in worse model performance.
It may help to remove outliers prior to applying the oversampling procedure, and this might be a helpful heuristic to use more generally.
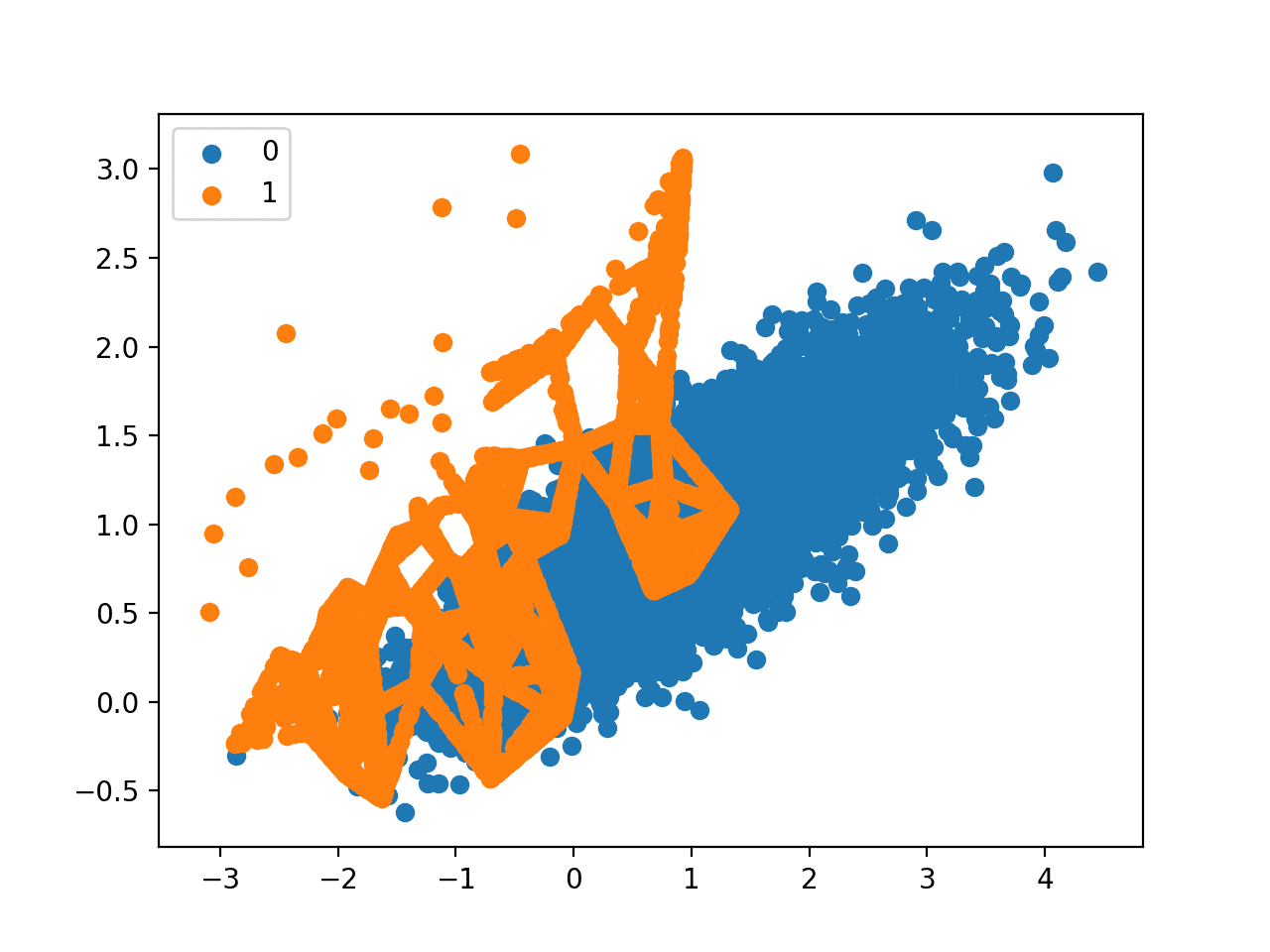
Scatter Plot of Imbalanced Dataset With Adaptive Synthetic Sampling (ADASYN)
Further Reading
This section provides more resources on the topic if you are looking to go deeper.
Books
- Learning from Imbalanced Data Sets, 2018.
- Imbalanced Learning: Foundations, Algorithms, and Applications, 2013.
Papers
- SMOTE: Synthetic Minority Over-sampling Technique, 2002.
- Borderline-SMOTE: A New Over-Sampling Method in Imbalanced Data Sets Learning, 2005.
- Borderline Over-sampling For Imbalanced Data Classification, 2009.
- ADASYN: Adaptive Synthetic Sampling Approach For Imbalanced Learning, 2008.
API
- imblearn.over_sampling.SMOTE API.
- imblearn.over_sampling.SMOTENC API.
- imblearn.over_sampling.BorderlineSMOTE API.
- imblearn.over_sampling.SVMSMOTE API.
- imblearn.over_sampling.ADASYN API.
Articles
Summary
In this tutorial, you discovered the SMOTE for oversampling imbalanced classification datasets.
Specifically, you learned:
- How the SMOTE synthesizes new examples for the minority class.
- How to correctly fit and evaluate machine learning models on SMOTE-transformed training datasets.
- How to use extensions of the SMOTE that generate synthetic examples along the class decision boundary.
Do you have any questions?
Ask your questions in the comments below and I will do my best to answer.







Hi
print(‘Mean ROC AUC: %.3f’ % mean(scores))
For calculatng ROC AUC, the examples make use of the mean function an not roc_auc_score, why?
Thanks
The ROC AUC scores are calculated automatically via the cross-validation process in scikit-learn.
Hi Jason,
Is there any way to use smote for multilabel problem.
Yes, you must specify to the smote config which are the positive/negative clasess and how much to oversample them.
Hello sir! I have 4 classes in my dataset (None(2552),Infection(2555),Ischemia(227),Both(621))..How can I apply this technique to my dataset?
Hi Emily…Hopefully the following will provide more clarity:
https://machinelearningmastery.com/multi-class-imbalanced-classification/
Hi Jason,
thanks for sharing machine learning knowledge.
How to get predictions on a holdout data test after getting best results of a classifier by SMOTE oversampling?
Best regards!
Mamadou.
Call model.predict() as per normal.
Recall SMOTE is only applied to the training set when your model is fit.
Hi Jason,
As you said, SMOTE is applied to training only, won’t that affect the accuracy of the test set?
Yes, the model will have a better idea of the boundary and perform better on the test set – at least on some datasets.
Just a clarifying question: As per what Akil mentioned above, and the code below, i am trying to understand if the SMOTE is NOT being applied to validation data (during CV) if the model is defined within a pipeline and it is being applied even on the validation data if I use oversampke.fit_resample(X, y). I want to make sure if it’s working as expected.
I saw a drastic difference in say, accuracy when I ran SMOTE with and without pipeline.
# define pipeline
steps = [(‘over’, SMOTE()), (‘model’, DecisionTreeClassifier())]
pipeline = Pipeline(steps=steps)
# evaluate pipeline
cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1)
scores = cross_val_score(pipeline, X, y, scoring=’roc_auc’, cv=cv, n_jobs=-1)
print(‘Mean ROC AUC: %.3f’ % mean(scores))
SMOTE is only applied on the training set, even when used in a pipeline, even when evaluated via cross-validation.
Makes sense! Like our fellow commenters mentioned, even in my case, train and validation have close accuracy metric but there is 7-8% dip for test set. What can be done to improve the performance of the test set (sorry for re-asking)?
P.S:
Just to be clear again, in my case – 3-class problem:
What I define as X_train is used to fit and evaluate the skill of the model . What happens under the hood is a 5-fold CV meaning the X_train is again split in 80:20 for five times where 20% of the data set is where SMOTE isn’t applied. This is my understanding.
Try the list of techniques here to improve model performance:
https://machinelearningmastery.com/framework-for-imbalanced-classification-projects/
And here:
https://machinelearningmastery.com/machine-learning-performance-improvement-cheat-sheet/
You say in a few different places something along the lines “SMOTE is only applied on the training set, even when used in a pipeline, even when evaluated via cross-validation.”
But your code:
steps = [(‘over’, SMOTE()), (‘model’, DecisionTreeClassifier())]
pipeline = Pipeline(steps=steps)
cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1)
scores = cross_val_score(pipeline, X, y, scoring=’roc_auc’, cv=cv, n_jobs=-1)
From what I am seeing, and what I see others commenting on, X & y get fed into the pipeline and go through cross validation as is. There is no training and test in this example broken out before the pipeline is applied. And your X & y go through cross validation with the oversampled data – so each fold is using that oversampled data for both the training and test So, even though you say you should only apply it to training data, your model is being optimized with SMOTE being applied to everything, which will inflate your metric results. Which of course, is what people are indicating is happening to their results, and their test results are significantly lower. My understanding is that you will want to cut into each fold and apply SMOTE only to the training data within the fold, which I do not see being done here.
In addition, you use ROC AUC as the metric to optimize for with imbalanced classification. This is not ideal, better to use something that focuses on the positive class, like precision-recall curve AUC, or average_precision.
Thank you for the feedback Keith! How has your recommendations performed in practice?
Hi !
SMOTE works for imbalanced image datasets too ?
Best Regards;
No, it is designed for tabular data.
You might be able to use image augmentation in the same manner.
Yours books and blog help me a lot ! Thank you very much !
Thanks, I’m happy to hear that!
Hello, I used image augmentation for my imbalanced image dataset but I still have low results for some classes so that influences the performance of my model. Also you have to know that I used it for all my data, I mean that I didn’t specify the classes with low images, I applied it for all my data. Can you help me with this? Thank you so much
Perhaps the model requires tuning, some of these suggestions will help:
https://machinelearningmastery.com/start-here/#better
Hi Jason, thanks for another series of excellent tutorials. I have encountered an error when running
X, y = pipeline.fit_resample(X, y)
on my own X & y imbalanced data. The error is :
“ValueError: The specified ratio required to remove samples from the minority class while trying to generate new samples. Please increase the ratio.”
from _validation.py”, line 362, in _sampling_strategy_float in imblearn/utils in the library.
Could you or anyone else shed some light on this error?
Thanks.
as a followup it seems I’ve not understood how SMOTE and undersampling function.
My input data size is:
{0: 23558, 1: 8466}
so a little under 1:3 for minority:majority examples of the classes
Now I understand I had the ratios for SMOTE() and RandomUnderSampler() “sampling_strategy” incorrect.
Onwards and upwards!
Happy to hear that, nice work!
Hi
When used with a gridsearchcv, does Smote apply the oversampling to whole train set or does it disregard the validation set?
You can use it as part of a Pipeline to ensure that SMOTE is only applied to the training dataset, not val or test.
Hi Jason,
Nice blog! And nice depth on variations on SMOTE. I was wondering:
Why do you first oversample with SMOTE and then undersample the majority class afterwards in your pipelines? Wouldn’t it be more effective the other way around?
Thanks!
It is an approach that has worked well for me. Perhaps try the reverse on your dataset and compare the results.
Hi Jason,
I’ve been perusing through your extremely helpful articles on imbalanced classification for days now. Thank you for providing such valuable knowledge to the machine learning community!
I had a question regarding the consequences of applying SMOTE only to the train set. If we apply SMOTE only to the train set but not to validation set or test set, the three sets will not be stratified. For example, if the train set transformed to a 50:50 distribution for class 1 and class 2, validation and test sets still maintain their original distribution 10:90, let’s say. Is this not a concern at all since we just care about baking the highest-performing MODEL which will be based only on the train set? If we apply SMOTE to only the train set wouldn’t the model also assume that the real-world data also assumes a 50:50 distribution between class 1 and class 2?
Thanks for your help in advance!
Thank you for your support!
No, you would stratify the split of the data before resampling. Then use a metric (not accuracy) that effectively evaluates the capability of natural looking data (val and test sets).
This is critical. Changing the nature of test and val sets would make the test harness invalid.
Confirm you have examples of both classes in the y.
Thank you for your tutorial.
I’d like to ask several things.
1. Could I apply this sampling techniques to image data?
2. After making balanced data with these thechniques, Could I use not machine learning algorithms but deep learning algorithms such as CNN?
Yes, but it is called data augmentation and works a little differently:
https://machinelearningmastery.com/how-to-configure-image-data-augmentation-when-training-deep-learning-neural-networks/
Yes, this tutorial will show you how:
https://machinelearningmastery.com/cost-sensitive-neural-network-for-imbalanced-classification/
Thank you for your answer.
I’ve used data augmentation technique once. So I can a little understand differency between data augmentation and oversampling like SMOTE.
In fact, I’d like to find other method except data augmentation to improve model’s performance. So, I wanted to try oversampling.
But, as follow as I understand as your answer, I can’t use oversampling such as SMOTE at image data . Am I right to understand?
Thank you again for your kind answer.
Correct, SMOTE does not make sense for image data, at least off the cuff.
Here are ideas for improving model performance:
https://machinelearningmastery.com/start-here/#better
Hello Jason,
In your ML cheat sheet you have advice to invent more data if you have not enough. Can you suggest methods or libraries which are good fit to do that?
Imblearn seams to be a good way to balance data. What about if you wish to increase the entire dataset size as to have more samples and potentially improve model?
SMOTE can be used.
Feature engineering is the more general approach:
https://machinelearningmastery.com/discover-feature-engineering-how-to-engineer-features-and-how-to-get-good-at-it/
Thank you for the great tutorial, as always super detailed and helpfull.
I’m working throught the wine quality dataset(white) and decided to use SMOTE on Output feature balances are below.
{6: 2198, 5: 1457, 7: 880, 8: 175, 4: 163, 3: 20, 9: 5}
In your opinion would it be possible to apply SMOTE in this multiclass problem?
I’ve managed to use a Regression model (KNN) that I belive does the task well but interested to get your take how to deal with similar class imbalance on multilclass problems as above?
Yes, SMOTE can be used for multi-class, but you must specify the positive and negative classes.
What does positive and negative means for multi-class? Based on the problem/domain, it can vary but let’s say if I identify which classes are positive and which are negative, what next?
You can apply SMOTE directly fir multi-class, or you can specify the preferred balance of the classes to SMOTE.
Also see an example here:
https://machinelearningmastery.com/multi-class-imbalanced-classification/
Thanks for sharing Jason.
In imblearn.pipeline the predict method says tahar it applies transforms AND sampling and then the final predict of the estimator.
Therefore isnt that a problem in crossvalscore the sampling will be applied on each validation sets ?
Thanks
Sorry, I don’t follow your question. Can you please rephrase or elaborate?
I think your description of borderline SMOTE1 and SMOTE2 is incorrect? To my knowledge SMOTE1 generates synthetic samples between the primary positive sample and some of the positive NNs and SMOTE2 also generates synthetic samples between the primary positive sample and some of the negative NNs (where the synthetics samples are closer to the primary positive sample). So negative samples are not generated.
Apologies if I am mistaken, love your content.
That description is quoted from the ICIC2005 paper.
you mentioned that : ” As in the previous section, we will first oversample the minority class with SMOTE to about a 1:10 ratio, then undersample the majority class to achieve about a 1:2 ratio.”
why? what is the idea behind this operation and why does this operation can inprove the performance.
This approach can be effective. It is important to try a range of approaches on your dataset to see what works best.
Sir Jason,
Can we use the above code for images
No, you would use data augmentation:
https://machinelearningmastery.com/how-to-configure-image-data-augmentation-when-training-deep-learning-neural-networks/
Hello Jason, thanks for the tutorial.
When using the lines:
# define pipeline
steps = [(‘over’, SMOTE()), (‘model’, RandomForestClassifier(n_estimators=100, criterion=’gini’, max_depth=None, random_state=1))]
pipeline = Pipeline(steps=steps)
# evaluate pipeline
cv = RepeatedStratifiedKFold(n_splits=5, n_repeats=2, random_state=1)
acc = cross_val_score(pipeline, X_new, Y, scoring=’accuracy’, cv=cv, n_jobs=-1)
I assume the SMOTE is performed for each cross validation split, therefore there is no data leaking, am I correct? Thank you
Correct. That is why we use pipelines.
Hello Jason,
Thank you for the post. I have some questions. My dataset consists NaN values and I am not allowed to drop them due to less no. of records. If I impute values with mean or median before splitting data or cross validation, there will be information leakage. To solve that problem, I need to use pipeline including SMOT and a model, and need to apply cross validation. Now, my question is, what if I have huge data set and I want to apply feature engineering (PCA or RFE) and want to explore all the steps step by step? If I define every steps in pipeline, how can I explore, where is the real problem in which method? Also I need more computation power to do trial and error methods on huge dataset. What is your suggestion for that?
My second question is, that I do not understand SMOT that you defined initially.
” SMOTE first selects a minority class instance a at random and finds its k nearest minority class neighbors. The synthetic instance is then created by choosing one of the k nearest neighbors b at random and connecting a and b to form a line segment in the feature space. The synthetic instances are generated as a convex combination of the two chosen instances a and b. ”
I couldn’t imagine what you want to say. Because of that I did not understand borderline SMOT as well. Could you please rephrase it and if possible could you please explain it with a small example?
Thank you in advance.
You must fit the imputer on the train set and apply to train and test within cv, a pipeline will help.
You can also step the k-fold cv manually and implement the pipeline manually – this might be preferred to you can keep track of what changes are made and any issues that might occur.
SMOTE works by drawing lines between close examples in feature space and picking a random point on the line as the new instance.
I hope that helps.
Hi! A quick question, SMOTE should be applied before or after data preparation (like Standardization for example) ? Or it’s irrelevant?
Thank you!
Probably after.
It is doing a knn, so data should be scaled first.
How to use SMOTE or any other technique related with SMOTE such as ADASYN, Borderline SMOTE, when a dataset has classes with only a few instances?
Some of the classes in my dataset has only 1 instance & some have 2 instances. When using these SMOTE techniques I get the error ‘Expected n_neighbors <= n_samples, but n_samples = 2, n_neighbors = 6'.
Is there any way to overcome this error? With RandomOversampling the code works fine..but it doesn't seem to give a good performance. And I'm unable to all the SMOTE based oversampling techniques due to this error.
I don’t think modeling a problem with one instance or a few instances of a class is appropriate.
Perhaps collect more data?
Perhaps delete the underrepresented classes?
Perhaps reframe the problem?
Hello I did tuning of smote parameters( k,sampling strategy) and took roc_auc as scoring on training data but how along with cross val score my model is evaluated on testing data (that ideally should not be the one on which smote should apply)
can you help me with how to apply best model on testing data(code required)
#Using Decsion Tree
Xtrain1=Xtrain.copy()
ytrain1=ytrain.copy()
k_val=[i for i in range(2,9)]
p_proportion=[i for i in np.arange(0.2,0.5,0.1)]
k_n=[]
proportion=[]
score_m=[]
score_var=[]
modell=[]
for k in k_val:
for p in p_proportion:
oversample=SMOTE(sampling_strategy=p,k_neighbors=k,random_state=1)
Xtrain1,ytrain1=oversample.fit_resample(Xtrain,ytrain)
model=DecisionTreeClassifier()
cv=RepeatedStratifiedKFold(n_splits=10,n_repeats=3,random_state=1)
scores=cross_val_score(model,X1,y1,scoring=’roc_auc’,cv=cv,n_jobs=-1)
k_n.append(k)
proportion.append(p)
score_m.append(np.mean(scores))
score_var.append(np.var(scores))
modell.append(‘DecisionTreeClassifier’)
scorer=pd.DataFrame({‘model’:modell,’k’:k_n,’proportion’:proportion,’scores’:score_m,’score_var’:score_var})
print(scorer)
models.append(model)
models_score.append(scorer[scorer[‘scores’]==max(scorer[‘scores’])].values[0])
models_var.append(scorer[scorer[‘score_var’]==min(scorer[‘score_var’])].values[0])
This is a common question that I answer here:
https://machinelearningmastery.com/faq/single-faq/can-you-read-review-or-debug-my-code
Hey Jason,
What kind of an approach can we use to over-sample time series data?
Good question, I hope I can cover that topic in the future.
Hello Jason,
Do you currently have any ideas on how to oversample time series data off the top of your head? I’d like to do some research/experiment on it in the meantime.Thank you!
No, in general I rather make recommendations after doing my homework.
Thank you very much!
You’re welcome.
did you have a chance to write about this topic(oversampling for time series data)?
Thanks for your suggestion. We will consider that.
Hello Jason,
I am here again reading your articles like I always did. By any chance did you write an article on time series data oversampling/downsampling?
I have been trying to find a manner to deal with time series data oversampling/ undersampling, but couldn’t find a proper manner yet to apply to this problem…
Hi Cel…You may find the following of interest:
https://web.cs.dal.ca/~branco/PDFfiles/c3.pdf
Hi Jason Brownie,
Thank you for the great description over handling imbalanced datasets using SMOTE and its alternative methods. I know that SMOTE is only for multi Class Dataset but I am curious to know if you have any idea of of using SMOTE for multi label Datasets?? or Do you have any other method or ideas apart from SMOTE in order to handle imbalanced multi label datasets.
Great question!
I’m not aware of an approach off hand for multi-label, perhaps check the literature?
I was working on a dataset as a part of my master thesis and it is highly imbalanced. So I tried experimenting directly using OnevsRestClassifier(without any oversampling) and naturally the classsifer gave worst results(the target value with high number of occurences is being predicted). So I tried testing with Random forest classifier taking each target column one at a time and oversampled with a randomsampler class which gave decent results after oversampling. And I am not sure if I can do it in this way.
Perhaps the suggestions here will help:
https://machinelearningmastery.com/framework-for-imbalanced-classification-projects/
I also found this solution.
https://github.com/scikit-learn-contrib/imbalanced-learn/issues/340
Nice.
Hi! Thanks for the great tutorial. Can SMOTE be used with 1. high dimensional embeddings for text representation? if so, what is any preprocessing/dimensionality reduction required before applying SMOTE?
Not sure off the cuff, perhaps experiment to see if this makes sense.
hi Jason , I am having 3 input Text columns out of 2 are categorical and 1 is unstructured text. Can you please help me how to do sampling. Output column is categorical and is imbalanced.
Perhaps use a label or one hot encoding for the categorical inputs and a bag of words for the text data.
You can see many examples on the blog, try searching.
I have used Pipeline and columntransformer to pass multiplecolumns as X but for sampling I ma not to find any example.For single column I ma able to use SMOTE but how to pass more than in X?
You may have to experiment, perhaps different smote instances, perhaps run the pipeline manually, etc.
Hi,
SMOTE requires 6 examples of each class.
I have a dataset if 30 class 0, and 1 class 1 .
Please advise if any solution.
Thank you
Perhaps try and get more examples from the minority class?
Perhaps try alternate techniques listed here:
https://machinelearningmastery.com/framework-for-imbalanced-classification-projects/
Hello Jason,
Many thanks for this article. I found it very interesting.
How can one apply the same ratio of oversampling (1:10) followed by under-sampling (1:2) in a pipeline when there are 3 classes?
The sampling strategy cannot be set to float for multi-class. What would you recommend?
Thank you.
John
Thanks.
First step is to group classes into positive and negative, then apply the sampling.
Could you shed some light on how one could leverage the parameter sampling_strategy in SMOTE?
Yes, what would you like to know exactly?
Hi Jason,
Thank you very much for this article, it’s so helpful (as always).
I have an inquiry:
Now my data are highly imbalanced (99.5%:0.05%). I am having over than 40,000 samples with multiple features (36) for my classification problem. I oversampled with SMOTE to have balanced data, but the classifier is getting highly biased toward the oversampled data. I assumed that its because of the “sampling_strategy”. So I tried {0.25, 0.5, 0.75,1} for the “sampling_strategy”. Its either getting highly biased towards the abundant or the rare class.
What do you think could be the problem?
SMOTE is not the best solution for all imbalanced datasets.
Perhaps try and compare alternative solutions:
https://machinelearningmastery.com/framework-for-imbalanced-classification-projects/
please tell me how i can apply two balancing technique first SMOTE and then one class learning algorithm on same dataset for better result
You can apply smote to the training set, then apply the one class classifier directly.
I don’t expect it would be beneficial to combine these two methods.
sir then what should i try for the best result by using smote and one more algo which makes an hybrid approch to handle imbalanced data.
Use trial and error to discover what works well/best for your dataset.
Hi Jason,
First, thanks for your material, it’s of great value!
I have a supervised classification problem with unbalanced class to predict (Event = 1/100 Non Event).
I have the intuition that using resampling methods such as SMOTE (or down/up/ROSE) with Naive Bayes models affect prior probabilities and such lead to lower performance when applied on test set.
Is that correct?
Thanks.
You’re welcome!
Yes.
Hi Dr.
Could SMOTE be applied to data that will be used for feeding an LSTM? (Since the order matters, it can interfere with the data right?)
Thanks in advance!
No, it is for tabular data only.
First of all, thanks for the response. Sorry, i think i don’t understand. Maybe I am wrong, but SMOTE could be applied to tabular data, before the transformation into sliding windows. Even in this case is not recommend to apply SMOTE ?
Thanks!
Perhaps, but I suspect data generation methods that are time-series-aware would perform much better.
Thanks you, Jason. Would you be able to point out an example of those time-series aware data generation methods?
Not off hand sorry. Perhaps try searching on scholar.google.com
hello sir how can we handle unbalanced dataset for lstm i have a csv file can we use smote technique or data generation could send me a link how we use oversampling cuz i have 3d array lstm input thank you so much
Hi Said…Please refer to the following:
https://machinelearningmastery.com/random-oversampling-and-undersampling-for-imbalanced-classification/
Jason,
I have a highly imbalanced binary (yes/no) classification dataset. The dataset currently has appx 0.008% ‘yes’.
I need to balance the dataset using SMOTE.
I came across 2 method to deal with the imbalance. The following steps after I have run MinMaxScaler on the variables
from imblearn.pipeline import Pipeline
oversample = SMOTE(sampling_strategy = 0.1, random_state=42)
undersample = RandomUnderSampler(sampling_strategy=0.5, random_state=42)
steps = [(‘o’, oversample), (‘u’, undersample)]
pipeline = Pipeline(steps=steps)
x_scaled_s, y_s = pipeline.fit_resample(X_scaled, y)
This results in a reduction in the size of the dataset from 2.4million rows to 732000 rows And the imbalance improves from 0.008% to 33.33%
While this approach
sm = SMOTE(random_state=42)
X_sm , y_sm = sm.fit_sample(X_scaled, y)
This increases the number of rows from 2.4million rows to 4.8 million rows and the imbalance is now 50%.
After these steps I need to split data into Train Test datasets….
What is the right approach here?
What factors do I need to consider before I choose any of these methods?
Should I run the X_test, y_test on unsampled data. This would mean, I split the data and do upsampling/undersampling only on the train data.
Thanks again.
No, the sampling is applied on the training dataset only, not the test set. E.g. split first then sample.
Hello Jason, Great article. One Issue i am facing while using SMOTE-NC for categorical data. I have only feature for categorization.
from imblearn.over_sampling import SMOTE
from imblearn.over_sampling import SMOTENC
sm = SMOTENC(random_state=27,categorical_features=[0,])
X_new = np.array(X_train.values.tolist())
Y_new = np.array(y_train.values.tolist())
print(X_new.shape) # (10500,)
print(Y_new.shape) # (10500,)
X_new = np.reshape(X_new, (-1, 1)) # SMOTE require 2-D Array, Hence changing the shape of X_mew
print(X_new.shape) # (10500, 1)
sm.fit_sample(X_new, Y_new)
But i am getting Error:
ValueError: Found array with 0 feature(s) (shape=(10500, 0)) while a minimum of 1 is required.
Can you please suggest how to deal with SMOTE if there is only one feature ?
Interesting, I wonder if it is a bug in smote-nc?
Perhaps try duplicating the column and whether it makes a difference?
sir how SMOTE can be applied on CSV file data
Load the data as per normal:
https://machinelearningmastery.com/load-machine-learning-data-python/
Then apply smote.
What is the criteria to UnderSample the majority class and Upsample the minority class.
OR
What is the criteria to Upsample the minority class only.
I don’t approach it that way. I think it’s misleading and intractable.
Instead, I recommend do the experiment and use it if it results in better performance.
SIR PLEASE PROVIDE TUTORIAL ON TEST TIME AUGMENTATION FOR NUMERICAL DATA
No problem, I have one written and scheduled to appear next week.
Sir is we apply feature selection technique first or data augmentation first.
Feature selection first would be my first thought.
Why are we implementing SMOTE on whole dataset “X, y = oversample.fit_resample(X, y)”? We should apply oversampling only on the training set. Am i right? What should be done to implement oversampling only on the training set and we also want to use stratified approach?
Correct, and we do that later in the tutorial when evaluating models.
In the first example I am getting you used to the API and the affect of the method.
Can you please refer that tutorial to me where we we are implementing smote on taining data only and evaluating the model? I also want to know that RepeatedStratifiedKfold works only on the training dataset only.
Yes the section “SMOTE for Classification” in the above tutorial uses a pipeline to ensure SMOTE is only applied on training data.
If you are new to using pipelines, see this:
https://machinelearningmastery.com/data-preparation-without-data-leakage/
cross_val_score oversample the data of training set only and do not oversample the training data. am i right?
When using a pipeline the transform is only applied to the training dataset, which is correct.
Thank You very much.
You’re welcome.
Hi Jason!
Thank you for such a great post!
I am working with an imbalanced data set (500:1). I want to get the best recall performance and I have tried with several classification algorithms, hyper parameters, and Over/Under sampling techniques. I will try SMOTE now !!!
From the last question, I understand that using CV and pipelines you oversample only the training set, right?
I have another question. My imbalanced data set is about 5 million records from 11 months. It is not a time series. I used data from the first ten months for training, and data from the eleventh month for testing in order to explain it easier to my users, but I feel that it is not correct, and I guess I should use a random test split from the entire data set. Is this correct?
You’re welcome.
Correct. Use a pipeline to only oversample the training set.
My best advice is to evaluate candidate models under the same conditions you expect to use them. If there is a temporal element to your data and how you expect to use the model in the future, try and capture that in your test harness.
I hope that helps.
Here are more ideas:
https://machinelearningmastery.com/framework-for-imbalanced-classification-projects/
Thank you
Hi Jason,
I followed your ideas at:
https://machinelearningmastery.com/framework-for-imbalanced-classification-projects/
I tried oversampling with SMOTE, but my computer just can’t handle it.
Then I tried using Decision Trees and XGB for imbalanced data sets after reading your posts:
https://machinelearningmastery.com/cost-sensitive-decision-trees-for-imbalanced-classification/
https://machinelearningmastery.com/xgboost-for-imbalanced-classification/
but I still get low values for recall.
I am doing random undersample so I have 1:1 class relationship and my computer can manage it. Then I am doing XGB/Decision trees varying max_depth and varying weight to give more importance to the positive class. My assumption is that I won’t overfit the model as soon as I use CV with several folds and iterations. Is that right?
Thanks
Well don on your progress!
Perhaps. Assumptions can lead to poor results, test everything you can think of.
Thank you
You’re welcome.
Hi Jason, excellent explanations on SMOTE, very easy to understand and with tons of examples!
I tried to download the free mini-course on Imbalance Classification, and I didn’t receive the PDF file.
May I please ask for your help with this? Thanks in advance!
Thanks.
Sorry to hear that, contact me directly and I will email it to you:
https://machinelearningmastery.com/contact/
Thank you, will do that!
You’re welcome.
Hi Jason, thanks for this tutorial it’s so useful as usual,
I have one inquiry, I have intuition that SMOTE performs bad on dataset with high dimensionality i.e when we have many features in our dataset. Is it true ?
Hmmm, that would be my intuition too, but always test. Intuitions breakdown in high dimensions, or with ml in general. Test everything.
Hi
When used with a gridsearchcv, does Smote apply the oversampling to whole train set or does it disregard the validation set?
You can use it as part of a Pipeline to ensure that SMOTE is only applied to the training dataset, not val or test.
hi jason,
do you mean if i use it in a imblearn’s own Pipeline class, it would be enough? no need for any parameter?
pipe = Pipeline(steps=[(‘coltrans’, coltrans),
(‘scl’,StandardScaler()),
(‘smote’, SMOTE(random_state=42))
]
)
X_smote,y_smote=pipe.fit_resample(X_train,y_train)
Yes.
Hi Jason,
Thanks for sharing. It really helps in my work 🙂
Let’s say you train a pipeline using a train dataset and it has 3 steps: MinMaxScaler, SMOTE and LogisticRegression.
Can you use the same pipeline to preprocess test data ?
Or should you have a different pipleine without smote for test data ?
How does pipeline.predict(X_test) that it should not execute SMOTE ?
Thanks.
The pipeline is fit and then the pipeline can be used to make predictions on new data.
Yes, call pipeline.predict() to ensure the data is prepared correctly prior to being passed to the model.
More on this here:
https://machinelearningmastery.com/data-preparation-without-data-leakage/
Hi Jason, SMOTE sampling is done before / after data cleaning or pre-processing or feature engineering??? I just want to know when we should do SMOTE sampling and why??
It depends on what data prep you are doing.
Probably after.
Hi, great article! I think there is a typo in section “SMOTE for Balancing Data”: “the large mass of points that belong to the minority class (blue)” –> should be majority I guess
Thanks! Fixed.
https://stackoverflow.com/questions/58825053/smote-function-not-working-in-make-pipeline
Sorry, I don’t have the capacity to read off site stackoverflow questions:
https://machinelearningmastery.com/faq/single-faq/can-you-comment-on-my-stackoverflow-question
Hi Jason,
TypeError: All intermediate steps should be transformers and implement fit and transform or be the string ‘passthrough’ ‘SMOTE(k_neighbors=5, n_jobs=None, random_state=None, sampling_strategy=’auto’)’ (type ) doesn’t
I get this error when running GridSearchCV. What is wrong?
Perhaps confirm the content of your pipeline ends with a predictive model.
Hi Jason,
if all my predictors are binary, can I still use SMOTE? seems SMOTE only works for predictors are numeric? Are there any methods other than random undersampling or over sampling? Thanks
Great question, I believe you can use an extension of SMOTE for categorical inputs called SMOTE-NC:
https://imbalanced-learn.readthedocs.io/en/stable/generated/imblearn.over_sampling.SMOTENC.html
Brilliant post Jason!
I wonder if we upsampled the minority class from 100 to 9,900 with a bootstrap (with replacement of course), whether we would get similar results than SMOTE … I put on my to-do list.
Thanks!
Probably not, as we are generating entirely new samples with SMOTE. Nevertheless, run the experiment and compare the results!
Interesting …. I will. Thanks Jason!
You’re welcome.
Hi, Jason
Thank you for your great article. It is really informative as always. Recently I read an article about the classification of a multiclass and imbalanced dataset. They used SMOTE for both training and test set and I think it was not a correct methodology and the test dataset should not be manipulated. please tell me if I am wrong and would you recommend a reference about the drawbacks and challenges of using SMOTE?
Thank you
Thanks!
Agreed, it is invalid to use SMOTE on the test set.
Hi Jason
Q1. Do we apply SMOTE on the train set after doing train/ test split?
Guess, doing SMOTE first, then splitting, may result in data leak as same instances may be present in both test and test sets.
Q2. I understand why SMOTE is better instead of random oversampling minority class. But say for a class imbalance of 1:100, why not just random undersample majority class? Not sure how SMOTE helps here !
Thanks
Vivek
Yes. Training set only.
Try many methods and discover what works best for your dataset.
Hi Jason,
What if you have an unbalanced dataset that matches the realistic class distribution in production. Say Class A has 1000 rows, Class B 400 and Class C with 60. What are the negative effects of having an unbalanced dataset like this. Say I use a classifier like Naive Bayes and since prior probability is important then by oversampling Class C I mess up the prior probability and stray farther away from the realistic probabilities in production. Should I try and get more data or augment the data that I have while maintaining this unbalanced distribution or change the distribution by oversampling the minority classes?
Thanks
The negative effects would be poor predictive performance.
I recommend testing a suite of techniques in order to discover what works best for your specific dataset.
This framework will help:
https://machinelearningmastery.com/framework-for-imbalanced-classification-projects/
Hello,
Thanks for your work, it is really useful. I have a question about the combination of SMOTE and active learning.
I am trying to generate a dataset using active learning techniques. From a pool of unlabelled data I select the new points to label using the uncertainty in each iteration. My problem is that the classes repartition is imbalanced (1000:1), my current algorithm can’t find enough points in Yes class. Do you think I could use SMOTE to generate new points of Yes class?
I am thinking about using borderline-SMOTE to generate new points and then label them. How can I be sure that the new points are not going to be concentrated in a small region?
I am not sure if I have explained the problem well. I need to find the feasible zone using the labeller in a smart way because labelling is expensive. Can you give me any advice?
Thanks.
Daniel
Perhaps try it and see?
I do SMOTE on the whole dataset, then normalize the dataset. After that I applied cross_val_score. Is it right that in cross_val_score, SMOTE will resampling only training set Code is here:
oversample = SMOTE()
X, Y = oversample.fit_resample(X, Y)
normalized = StandardScaler()
normalized_X = normalized.fit_transform(X)
clf_entropy = DecisionTreeClassifier(random_state = 42)
y_pred = cross_val_predict(clf_entropy, normalized_X, Y, cv=15)
No. If you use a pipeline it will work as you describe.
Hi Jason .
Thanks for your post. I have two Qs regards SMOTE + undersampling example above.
“under = RandomUnderSampler(sampling_strategy=0.5)” . Why would we undersample the majority class to have 1:2 ratio and not have an equal representation of both class?
2. If I were to have an imbalanced data such that minority class is 50% , wouldn’t I need to use PR curve AUC as a metric or f1 , instead of ROC AUC ?
“scores = cross_val_score(pipeline, X, y, scoring=’roc_auc’, cv=cv, n_jobs=-1)”
Thanks !!
It is a good idea to try a suite of different rebalancing ratios and see what works. I found this ratio on this dataset after some trial and error.
This will help you choose a metric:
https://machinelearningmastery.com/tour-of-evaluation-metrics-for-imbalanced-classification/
Thanks Jason. Applying the same now .
Thanks!
Jason , I am trying out the various balancing methods on imbalanced data . How ever , yet to feel convinced on how balancing the training data set will allow the algorithm learn and work fairly well on the imbalanced test data ? Is this then dependent on how good the features are ? Means , if I see that after various methods of balancing the train data set , the model does not generalise well on test data , I need to relook at the feature creation ??
Thanks!!
Hard to say, the best we can do is used controlled experiments to discover what works best for a given dataset.
Thanks !
You’re welcome.
Hi Jason,
Thank you so much for your explanation. I have a question when fitting the model with SMOTE:
Why you use .fit_resample instead of .fit_sample? What is the difference between the two functions?
Also, is there any way to know the index for original dataset after SMOTE oversampling? How can I know what data comes from the original dataset in the SMOTE upsampled dataset?
Thanks!
Sorry, the difference between he function is not clear from the API:
https://imbalanced-learn.readthedocs.io/en/stable/generated/imblearn.over_sampling.SMOTE.html
Perhaps experiment with both and compare the results.
Hi, I applied the SMOTE for Balancing Data Code, firstly, I had 27 features in my data, when I defined the dataset in make_classification, I wrote the n_features=27 instead of 2, Is It Correct? and Can I apply the SMOTE for Balancing Data when my goal from the model is Prediction?
Secondly, How can I save the new data set in a CSV?
Thanks!
If you have your own data, you don’t need to use make_classification as it is a function for creating a synthetic dataset.
Ok, I want to apply the SMOTE, my data contains 1,469 rows, the class label has Risk= 1219, NoRisk= 250, Imbalanced data, I want to apply the Oversampling (SMOTE) to let the data balanced.
firstly, I run this code that showed me diagram of the class label then I apllyied the SMOTE,
target_count = data[‘Having DRPs’].value_counts()
print(‘Class 1:’, target_count[1])
print(‘Class 0:’, target_count[0])
print(‘Proportion:’, round(target_count[1] / target_count[0], 2), ‘: 1′)
target_count.plot(kind=’bar’, title=’Count (Having DRPs)’);
****
(Over-sampling: SMOTE):
from imblearn.over_sampling import SMOTE
smote = SMOTE(ratio=’minority’)
X_sm, y_sm = smote.fit_sample(X, y)
plot_2d_space(X_sm, y_sm, ‘SMOTE over-sampling’)
It gave me an error:
TypeError: __init__() got an unexpected keyword argument ‘ratio’
How can I solve this issue!
Sorry to hear that, perhaps some of these tips will help:
https://machinelearningmastery.com/faq/single-faq/can-you-read-review-or-debug-my-code
Hi, Fatima! could you please explain to me how you did smote on your dataset? I have my dataset of 4 classes (infection(2555),none(2552),both(621),ischemia(227))
Hi Jason, I applied the SMOTE on my data and I solved the imbalanced data, the next step I want to start Deep Learning(DL), in DL Do I have to save the new data ( balanced ) and then start DL algorithms on the new data ??
Thanks!
Only the training set should be balanced, not the test set.
You can transform the data in memory before fitting your model. Or you can save it if that is easier for you.
Hello Fatima! how did you apply smote to your dataset? In my dataset, I have 4 classes (none (2552), ischemia (227), both (621), and infection (2555). How can I integrate SMOTE?
Dear Jason,
Thanks for the awesome article!
I tried to implement the SMOTE in my project, but the cross_val_score kept returning nan.
Then I tried your piece of code:
# decision tree evaluated on imbalanced dataset with SMOTE oversampling
from numpy import mean
from sklearn.datasets import make_classification
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import RepeatedStratifiedKFold
from sklearn.tree import DecisionTreeClassifier
from imblearn.pipeline import Pipeline
from imblearn.over_sampling import SMOTE
# define dataset
X, y = make_classification(n_samples=10000, n_features=2, n_redundant=0,
n_clusters_per_class=1, weights=[0.99], flip_y=0, random_state=1)
# define pipeline
steps = [(‘over’, SMOTE()), (‘model’, DecisionTreeClassifier())]
pipeline = Pipeline(steps=steps)
# evaluate pipeline
cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1)
scores = cross_val_score(pipeline, X, y, scoring=’roc_auc’, cv=cv, n_jobs=-1)
print(‘Mean ROC AUC: %.3f’ % mean(scores))
This also returned nan.
I can’t figure out why it returns nan. In you article you describe that you do get an answer for this code snippet.
Thanks a lot!
Samuel
That’s surprising, perhaps change the cv to raise an error on nan and inspect the results.
Even I keep getting the same error
Perhaps some of these tips will help:
https://machinelearningmastery.com/faq/single-faq/can-you-read-review-or-debug-my-code
Hi,
Even I keep getting nan values as scores. Were you guys able to resolve it? If so, can you please provide some tips?
from sklearn.model_selection import StratifiedKFold
from sklearn import metrics
cv = StratifiedKFold(n_splits=10,shuffle=True)
classifier = AdaBoostClassifier(n_estimators=200)
y = df['label'].values
X = df
X = X.drop('label',axis=1)
X = X.values
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.10, random_state = 0, stratify = y)
oversampler= sv.CCR()
X_samp, y_samp= oversampler.sample(X_train, y_train)
X_train = X_samp
y_train = y_samp
tprs = []
aucs = []
mean_fpr = np.linspace(0, 1, 100)
plt.figure(figsize=(10,10))
i = 0
# cv.sh
for train, test in cv.split(X_train, y_train):
probas_ = classifier.fit(X_train[train], y_train[train]).predict_proba(X_train[test])
# Compute ROC curve and area the curve
fpr, tpr, thresholds = metrics.roc_curve(y_train[test], probas_[:, 1])
tprs.append(np.interp(mean_fpr, fpr, tpr))
tprs[-1][0] = 0.0
roc_auc = metrics.auc(fpr, tpr)
aucs.append(roc_auc)
plt.plot(fpr, tpr, lw=1, alpha=0.3,
label='ROC fold %d (AUC = %0.2f)' % (i, roc_auc))
i += 1
plt.plot([0, 1], [0, 1], linestyle='--', lw=2, color='r',
label='Chance', alpha=.8)
mean_tpr = np.mean(tprs, axis=0)
mean_tpr[-1] = 1.0
mean_auc = metrics.auc(mean_fpr, mean_tpr)
std_auc = np.std(aucs)
plt.plot(mean_fpr, mean_tpr, color='b',
label=r'Mean ROC (AUC = %0.2f $\pm$ %0.2f)' % (mean_auc, std_auc),
lw=2, alpha=.8)
std_tpr = np.std(tprs, axis=0)
tprs_upper = np.minimum(mean_tpr + std_tpr, 1)
tprs_lower = np.maximum(mean_tpr - std_tpr, 0)
plt.fill_between(mean_fpr, tprs_lower, tprs_upper, color='grey', alpha=.2,
label=r'$\pm$ 1 std. dev.')
plt.xlim([-0.01, 1.01])
plt.ylim([-0.01, 1.01])
plt.xlabel('False Positive Rate',fontsize=18)
plt.ylabel('True Positive Rate',fontsize=18)
plt.title('Cross-Validation ROC of ADABOOST',fontsize=18)
plt.legend(loc="lower right", prop={'size': 15})
plt.show()
check this output :
https://ibb.co/PYLs8qF
i am confused cause smote after adaboost for train works good but the test set is not good.
https://ibb.co/yPSrLx2
edit : I have used CCR which is a variant of smote. also It is CCR+Adaboost
Well done!
hi jason, can i ask? i applied metode smote bagging svm and smote boosting svm but always eror, can u help me to found the coding in python?
I don’t have the capacity to debug your code sorry, perhaps these suggestions will help:
https://machinelearningmastery.com/faq/single-faq/can-you-read-review-or-debug-my-code
Hi @jasonBrowniee, Thanks for the above example. Quick Question, for SMOTE you have used over sampling followed by Random Under Sampling, wondering if we use ADASYN or SVMSMOTE do you suggest we use random under sampling as we do in case of SMOTE?
Perhaps try a few different combinations and discover what works well/best for your specific dataset.
Hi Jason,
I hope you are doing well! Is there a need to upsample with Smote() if I use Stratifiedkfold or RepeatedStratifiedkfold? I think that my stratified folding already takes care of class imbalance. So is there a situation where you would prefer Smote over Stratified folding?
Cheers
SMOTE can be used with or without stratified CV, they address different problems – sampling the training dataset vs evaluating the model.
I don’t believe this technique “actually” works in many cases. You can read Jonas Peters’ work to understand why. It is really an example of Machine Learning Hocus-Pocus, or the creative side of Data Science which defines “works” as “I tried it and saw an improvement” anecdotal evidence. It is bad overall to not rigorously evaluate such methods through analytical and logical approaches.
Thanks for sharing your thoughts Michael.
Hi Jason,
Thanks for all of these heuristic alternatives you suggested for balancing datasets.
You’re welcome.
Hi Dr Jason. I saw at few articles, authors were compared imbalanced class and overlapped class. Do you have an article for that?
Almost all classes overlap – if not the problem would be trivial (e.g. linearly separable).
Not sure what you mean exactly?
Thanks Dr.
I am actually new to ML and quite interested in imbalanced classification. While feeding my mind to understand the fundamentals of ML and imbalanced from here: https://machinelearningmastery.com/start-here/.
Then, I started read others just to strengthen and verify my understanding. I found this article: https://link.springer.com/chapter/10.1007/978-3-642-13059-5_22 telling the difference between imbalanced and overlap.
Maybe because of my fundamental is not really strong, I’m not really understand what they thought in this article. So, I came to your blog as usual (it really helps newbie like me), to find article that share about the different between overlap and imbalance. Unfortunately, I could not find any. ????
Thanks for sharing, I’m not familiar with the article sorry.
OK Dr. Jason
Btw, is it important for me to understand overlapping issue in dataset? An article for that, maybe?
Thanks once again Dr
I don’t think so, I’ve not heard of the concept before.
OK Dr, Thank you so much
Hi Jason thanks for this very informative post. But just wondering, does it make sense for me to tune the model hyperparameters on an over/undersampled data set, like this?
paramgrid_rf = {‘n_estimators’: [500],
‘max_depth’: [4],
‘random_state’: [0],
‘max_features’: [‘sqrt’],
‘criterion’ :[‘mse’]
}
rfc = GridSearchCV(RandomForestRegressor(), paramgrid, cv=5)
steps = [(‘over’, SMOTE(sampling_strategy=0.2)), (‘model’, rfc)]
pipeline = Pipeline(steps=steps)
# evaluate pipeline
cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1)
scores = cross_val_score(pipeline, x_train, y_train, scoring=’f1′, cv=cv, n_jobs=-1)
Perhaps.
Do anything you can to get better results on your test harness.
Please specify which modules are needed. Took me an hour to find the damn where attribute from numpy
This tutorial will show you how to setup your development environment:
https://machinelearningmastery.com/setup-python-environment-machine-learning-deep-learning-anaconda/
my above comment looks too negative. THIS IS AWESOME; just please specify which modules to import.
Thank you for your work
The complete code example at the end of each sections has the import statements with the code.
This will help you copy the code from the tutorial:
https://machinelearningmastery.com/faq/single-faq/how-do-i-copy-code-from-a-tutorial
Hi Jason,
That was a very useful tutorial.
Thank u.
You’re welcome!
thanks, but i confused with appling smote on training data or on x and y as in examples and what the difference between them
This will explain X and y:
https://machinelearningmastery.com/faq/single-faq/what-are-x-and-y-in-machine-learning
ok, that are x and y (feature and target ) but why you applying smote on it? is smote applying on the training data means x splits into train and test and y as it the applying smote on xtrain and ytrain
Yes, SMOTE is applied to the training dataset only.
The above example shows you how to use the SMOTE class and the effect it has – so you feel comfortable with it and can start using it on your own project.
Hi,Jason
Can we implement SMOTENC with FAMD(prince) in a imblearn pipeline? If yes can you provide me with some reference regarding the approach and code.
I don’t know off hand, sorry. Perhaps explore it with a prototype.
Thanks
Hi Jason, thanks for the great content of SMOTE. I have a categorical variable in my data which is location. I can use that in resampling thanks to SMOTENC. But is there a way to implement SMOTE so that I can obtain homogeneity with respect to the minority class in location. So SMOTE would generate synthetic data in locations that initially have low instances of the minority class.
You might need to implement the algorithm yourself to have such fine grained control over where the algorithm chooses to resample.
Thanks for your prompt reply, as always!
You’re welcome.
Can we apply SMOTE for testing dataset also?
No, SMOTe is only applied to the training dataset.
So how should I do if the testing data is imbalance? I split the date set into 70% training set and 30% testing set. After I use smote to balance training set and then I want to test the model on testing set,then AUC will very low due to the imbalance testing set ,how should I do?Thank you very much!
Perhaps AUC is not the best metric for your problem?
Perhaps you can use repeated k-fold cross-validation to estimate the AUC?
Hi Json,
This was very succinct article on imbalance class. Thanks a lot for the article and the links to original paper.
Thanks!
Hi, great article, but please do not recommend using sudo privileges when installing python packages from pip! You are basically giving admin privileges to some random script pulled from the internet which is really not good practice, and even dangerous. For more references, look here: https://askubuntu.com/a/802594
Thanks a lot!
Thanks for sharing.
Hello Jason
I’m newbie here. I’m dealing with time series forecasting regression problem. That’s mean the prediction model is required to learn from the series of past observations to predict the next value in the sequence.
I’m using the dataset 1998 World Cup Web site (consists of all the requests made to the 1998 World Cup Web site between April 30, 1998 and July 26, 1998). Here the FTP link: ftp://ita.ee.lbl.gov/html/contrib/WorldCup.html
I preprocess the dataset by aggregating all logs that occur within the same minute into one accumulative record.
I want to ask if my dataset imbalanced? and Why?
Thanks for your help.
No. Typically imbalance is for classification tasks, and you said your problem is regression (predicting a numerical value).
Hello Jason, thanks for article.
I saw an article about SMOTE and I am confused. Here is the code they used:
from imblearn.over_sampling import SMOTE
sm = SMOTE(random_state=42)
X_sm, y_sm = sm.fit_resample(X, y)
X_train, X_test, y_train, y_test = train_test_split(
X_sm, y_sm, test_size=0.25, random_state=42
)
model = RandomForestClassifier(random_state=42)
model.fit(X_train, y_train)
preds = model.predict(X_test)
You said SMOTE is applied only on training set. So the code above is wrong?
And here is article in case you want to take a look:
https://towardsdatascience.com/how-to-effortlessly-handle-class-imbalance-with-python-and-smote-9b715ca8e5a7
I try not to comment on other peoples code – they can do whatever they like.
Yes. Fatally.
Hi, i’d like to thank you for your blog. It’s been really a great help for me. as a beginner, I’d like to ask you a question please. Does applying SMOTE with cross validation results in a biased model. I mean, when you set the pipeline to apply SMOTE then model fitting, does cross validation apply the validation process on the original test set or the over sampled test set? I saw on a stackoverflow post that when we use SMOTE it should be done only on the training set and the model should be tested only on the original data. Does cross validation meet this criteria too? Thanks.
When using SMOTE in a pipeline it is only applied to the training set, never the test set within a cross-validation evaluation/test harness.
Hi, first of all, I just wanna say thanks for your contribution. And i have a question
scores = cross_val_score(pipeline, X, y, scoring=’roc_auc’, cv=cv, n_jobs=-1)
score = mean(scores)
it’s seen mean nothing when you caculate your cross_val_score on your training data, I mean AUC is matter when you caculate on your testing data. I have high auc cross-validation but 0.5 on testing data.
Sorry, I don’t understand your question. Perhaps you could rephrase it?
Hello, thank you,
I would like toknow if smote method work for text data. I am dealing text classification with imbanlanced data.
No, tabular data only.
Thank you for your great post. I think SMOTE is only for the imbalanced tabular dataset, which is a classification problem. Do you know any augmentation methods for regression problems with a tabular dataset? Many thanks!
You’re welcome.
Correct.
No sorry. Most resampling methods are designed for imbalanced classification (not regression) as far as I have read.
Hope this makes sense, but I think the ROC scores in the CV as calculated is not right.
Here’s the reason: in your pipeline code there is over sampling and undersampling done. But I want the scores to be computed on the original dataset, not on the sample. If you generate synthetic then of course you can make the ROC look better on the dataset with synthetic data, but I want to know how well the dataset perform on the original data.
Currenlty the scores are
score = ROC(sampled(X), sampled(y))
but I want
score = ROC(X, y)
actually, I have removed the part about k-fold, but you can what I mean. So i think the code is not doing things correctly
ROC scores are only calculated using original data, no synthetic data. E.g. SMOTE is only used on train, not test.
The pipeline ensures this to be the case.
Hi Dr. Jason,
How do we apply SMOTE method to imbalanced classification time-series data? Also, is repeatedStratefied() applied to time series cv k-fold?
Thank you
SMOTE is not appropriate for time series.
Cross-validation is not appropriate for time series either, you must use methods like walk-forward validation:
https://machinelearningmastery.com/backtest-machine-learning-models-time-series-forecasting/
Hi Jason,
After balancing my severely imbalanced data (1:1000) using Smote, do I need to create an ensemble classifier in order to avoid overfitting with the minority class, due to oversampling of minority class and under sampling the majority class? Also if I used Random Forest which is an ensemble by itself, can I create an ensemble of random forests i.e. an ensemble of ensembles? Or would this lead to overfitting?
Thank you
Try it and compare results.
Focus on the evaluation metric. Overfitting is one possible cause of poor results.
Also, under-sampling and over-sampling may lead to loss of information from the dataset.
How do you compare this (Smote) to using weights for random forest instead?
Thank you.
Perhaps compare a RF directly with a RF fit on a SMOTE version of the dataset.
Hello Jason,
Thank you for the great description over handling imbalanced datasets using SMOTE and its alternative methods.
I have a small doubt when applying SMOTE followed by PCA. What is the best approach to apply SMOTE? Is it PCA first and then SMOTE or vice versa? What is the rationale behind this?
Perhaps try a few different approaches/orderings and discover what works best for your dataset and model.
Okay! Thanks Jason
Hello Jason, As always, Thank you for the wonderful article. I am working on a disease progression prediction problem. Objective is to predict the disease state (one of the target classes) at a future point in time, given the progression of the disease condition over the time (temporal dependencies in the progression).
Majority of my dataset belongs to “Healthy” condition and I have only few samples representing various other disease conditions (other target classes). Can you pls advise on how to oversample the minority class samples in this particular scenario?, Thanks again.
Perhaps try SMOTE described above and compare results to not using it?
Thank you Jason. Did you mean, compare between the results using SMOTE and results using ‘other’ techniques? Can you pls elaborate bit on it? thanks again.
Yes, perhaps other oversampling methods or no oversampling methods.
Hi Jason,
I have a question about the numbers on the axis of the scatterplot (-0,5 till 3 and -3 till 4) . What is de meaning of the axis values?
They are the values of the input variables, just a demonstration of what SMOTE does.
thnx, for your answer. I think I don’t understand it completely yet:
I thought the values were the classlabels (input = classlabeld). But ho do I interpret then the x-axis and y-axis?
thanks a lot again?
X is variable 1, y is variable 2, color is class label.
Should developers always calibrate predicted probabilities in combination with SMOTE or other rebalancing techniques?
Probably not – only when the model does not natively provide probabilities.
Hi , Jason , it is great article and it is really helping me understanding SMOTE . I have more question about K mean SMOTE and CURE SMOTE , may you add that 2 with example into your paper ? because i thin it is difficult to implement since not many example out there. Thanks in advance
Thanks for the suggestion.
How could I apply SMOTE to multivariate time series data like human activity dataset?
thanks
SMOTE would not be appropriate for time series or sequence data.
Perhaps you can check the literature for an oversampling method that is appropriate for time series data.
Thanks a lot!
Just one quick question.
In this sentence below
“This is referred to as Borderline-SMOTE1, whereas the oversampling of just the borderline cases in minority class is referred to as Borderline-SMOTE2.”
I guess 1 and 2 should be switched if 1 affects only one class, and 2 affects both classes.
“””Borderline-SMOTE2 not only generates synthetic examples from each example in DANGER and its positive nearest neighbors in P, but also does that from its nearest negative neighbor in N.”””
Please let me know if I’m getting it wrong.
Thanks!
Thanks.
# define pipeline
steps = [(‘over’, SMOTE()), (‘model’, DecisionTreeClassifier())]
pipeline = Pipeline(steps=steps)
# evaluate pipeline
cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1)
scores = cross_val_score(pipeline, X, y, scoring=’roc_auc’, cv=cv, n_jobs=-1)
Hi, Jason. Where is the part of splitting of the training and validation datasets before oversampling the training dataset only in each fold? I don’t get it. Is it implicit? I couldn’t be sure that oversampling process is applied only to the training dataset and not to validation dataset after splitting. Thanks.
Here you’re doing CV. The model to the CV is the pipeline, which includes SMOTE and a decision tree. It will do k-fold and feed the split into the model to train, the use the hold-out set to test. All are done inside RepeatedStratifiedKFold() function.
# define pipeline
steps = [(‘over’, SMOTE()), (‘model’, DecisionTreeClassifier())]
pipeline = Pipeline(steps=steps)
# evaluate pipeline
cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1)
scores = cross_val_score(pipeline, X, y, scoring=’roc_auc’, cv=cv, n_jobs=-1)
Hi, Jason. In here you are giving smote algorithm to the cross validation score directly. which means that it will first apply smote algorithm then split the dataset. So we also applied SMOTE to the fold that will be used to test. Shouldn’t we first do smote then give the dataset to cross_val_score to avoid this.
No. The pipeline will be fitted using the split dataset, not entire dataset. The ordering of steps like this is to avoid data leaking.
Well ı know realize that nothing is supposed to change when we do it like this but even so i tried to do it and to my surprise. ROC AUC score is increased on average 0.1 percent why does this happen ?
new ROC AUC scores
> k=1, Mean ROC AUC: 0.951
> k=2, Mean ROC AUC: 0.927
> k=3, Mean ROC AUC: 0.925
> k=4, Mean ROC AUC: 0.919
> k=5, Mean ROC AUC: 0.925
> k=6, Mean ROC AUC: 0.909
> k=7, Mean ROC AUC: 0.899
with the code from here
> k=1, Mean ROC AUC: 0.835
> k=2, Mean ROC AUC: 0.825
> k=3, Mean ROC AUC: 0.840
> k=4, Mean ROC AUC: 0.855
> k=5, Mean ROC AUC: 0.846
> k=6, Mean ROC AUC: 0.830
> k=7, Mean ROC AUC: 0.845
My code:
for k in k_values:
# define pipeline
model = DecisionTreeClassifier()
over = SMOTE(sampling_strategy=0.1, k_neighbors=k)
under = RandomUnderSampler(sampling_strategy=0.5)
steps = [(‘over’, over), (‘under’, under)]
pipeline = Pipeline(steps=steps)
X_t,y_t = pipeline.fit_resample(X,y)
# evaluate pipeline
cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1)
scores = cross_val_score(model, X_t, y_t, scoring=’roc_auc’, cv=cv, n_jobs=-1)
score = mean(scores)
print(‘> k=%d, Mean ROC AUC: %.3f’ % (k, score))
Are you doing right here? I see “pipeline” is defined but you evaluated “model”
Hello Jason I’m working on a data that is balanced, but have few instances, 171 instance for each class, please guide me on how to use smote and increase ( oversampling) each class, thanks.
I don’t see the imblearn library allows you to do that. But you can purposely add one fake majority class to the data and apply SMOTE. Only afterwards, you remove that fake class.
Hi Jason, Thanks for your fantastic website,
I’ve successfully installed imbalanced-lean, but when I’m trying to import imblearn, it gives me the following error:
AttributeError: partially initialized module ‘logging’ has no attribute ‘StreamHandler’ (most likely due to a circular import)
Do you know how I can fix that?
Thanks a ton in advance for your answer.
Hi Hosein…You may find the following of interest:
https://imbalanced-learn.org/stable/install.html
Hi Jason, Thank you for the clear and informative tutorials from all your posts. One question I have for these under/over sampling method or change weight method, don’t we need to scale back after the training phase like in the validation/test step? Thank you.
Hi Eva…It is recommended that data be inverse transformed when computing the performance metrics during validation.
Hi all. One question, please. When we use smote we result in class balanced. In that sense, we can use accuracy for our metric? Or again we must use something from here https://machinelearningmastery.com/tour-of-evaluation-metrics-for-imbalanced-classification/ ?
Hi Jason, I discovered your site yesterday and i´m amazed with your content.
I have a question:
for label, _ in counter.items():
row_ix = where(y == label)[0]
pyplot.scatter(X[row_ix, 0], X[row_ix, 1], label=str(label))
pyplot.legend()
pyplot.show()
How does this loop work ?
Thank you so much
Hi Tomas…My recommendation would be to implement such in your Python environment to best understand. In other words, experiment with it to learn more.
How do you convert convert the array back to a data frame especially if you want to pass it through features selection or ranking after you applied the SMOTE and Undersampling?
Hi John…You may find the following of interest:
https://github.com/scikit-learn-contrib/imbalanced-learn/issues/534
As Jason points out, the synthetic samples from SMOTE are convex combinations of original sample when the features are numerical. How does SMOTE address categorical features? A naive way is to use the dominant category in the component sample, but I haven’t seen it in any paper.
Thanks for the great explanation Jason.
Your blog is really awesome.
Thank you for the great feedback!
Thank you very much.
I tried to use it by “from imblearn.over_sampling import SMOTE”
But the python says:
No module named ‘imblearn’
So, I tried to install it. I used the following line
“!pip install -U imbalanced-learn”
Still, The same error.
What should I do?
I appreciate your time in advance
Hi Ali…You may wish to try to Google Colab while you are researching options to correct your local environment.
Great article and tutorial, thank you.
In my case, I have a 16/84 imbalanced dataset and did multiple tests with multiple estimators with and without SMOTE. I have overall better results (F1 and Matthew Corr) without SMOTE. Should I keep it imbalanced?
Hi Marc…I see no issue with your suggestion. Have you implemented your model? Please let us know your findings.
thank you for this tutorial. in the previous example of SMOTE, the data has two x features, what if the dataset has multiple x, and what do the x-axis and y-axis represent in scatter plot figures?
Hey James,
Why does using linear approximation for replicating the data points work in SMOTE? Why cant it be non linear?
Hi Shiraj…There are other suitable methods that could be used. Have you applied non-linear methods?
Hello James, thank u for your hardwork!
Do you know if SMOTE method can be apply for Unsupervised Learning (Clustering?
I’m trying to prepare my data to apply the clustering, and I can’t understand if I’m supposed to “correct” the imbalanced data in this case.
Hi Marta…You are very welcome! You may find the following resource helpful:
https://www.sciencedirect.com/science/article/abs/pii/S0020025521001985
Hello Jason,
Hope you will read this query.
Wanted to know how would one get final production model in case we use SMOTE?
Well, in general after cv once we are happy with results, we would combine all splits of data and apply/repeat procedure with best hyperparameters obtained from cv on entire data. In case of resampling – do we balance entire data to get production model?
Hi Nishant…You may find the following of interest:
https://machinelearningmastery.com/a-first-course-on-deploying-python-projects/
Thanks James, appreciate the link. Found it interesting but does not answer my query.
Hi,
Thank you so much for sharing your ML knowledge!
Let’s say we have an imbalanced dataset (1:100). Because the dataset should be stratified, three subsets of training, validation, and testing have the same ratio. Now, if we apply SMOTE only on the training subset to get a ratio of for example 1:2, the other two subsets remain in the same imbalanced 1:100 ratio.
Is my understanding correct?
So, I do not understand how SMOTE can increase performance while there is the mentioned issue.
Thank you,
Mohammad
Hi Mohammad…The following resource may be of interest in terms of when to apply SMOTE.
https://analyticsindiamag.com/how-can-smote-technique-improve-the-performance-of-weak-learners/
Thank you, James. I appreciate your response, but I did not find my response on that website.
Hi Jason, thank you for sharing the tutorial
Btw, can you explain how you apply the oversampling only to the training dataset? I don’t see the train-test splitting in your code examples
You are very welcome Abitama! The following resources may be of interest:
https://datascience.stackexchange.com/questions/61858/oversampling-undersampling-only-train-set-only-or-both-train-and-validation-set
https://machinelearningmastery.com/combine-oversampling-and-undersampling-for-imbalanced-classification/
!pip install scikit-learn
!pip install imblearn
from imblearn.over_sampling import SVMSMOTE
Shows error:
cannot import name ‘_MissingValues’ from ‘sklearn.utils._param_validation’ (C:\Users\Acer\anaconda3\Lib\site-packages\sklearn\utils\_param_validation.py)
I am unable to resolve the issue.
Please let me know how I can resolve this issue or alternate approach of using sklearn and imblearn
Hi Sasadhar…The following resource may be of interest to you:
https://stackoverflow.com/questions/76593906/how-to-resolve-cannot-import-name-missingvalues-from-sklearn-utils-param-v
Hi,
Do you have examples of SMOTE + GAN sampling?