Long Short-Term Memory (LSTM) models are a recurrent neural network capable of learning sequences of observations.
This may make them a network well suited to time series forecasting.
An issue with LSTMs is that they can easily overfit training data, reducing their predictive skill.
Weight regularization is a technique for imposing constraints (such as L1 or L2) on the weights within LSTM nodes. This has the effect of reducing overfitting and improving model performance.
In this tutorial, you will discover how to use weight regularization with LSTM networks and design experiments to test for its effectiveness for time series forecasting.
After completing this tutorial, you will know:
- How to design a robust test harness for evaluating LSTM networks for time series forecasting.
- How to design, execute, and interpret the results from using bias weight regularization with LSTMs.
- How to design, execute, and interpret the results from using input and recurrent weight regularization with LSTMs.
Kick-start your project with my new book Deep Learning for Time Series Forecasting, including step-by-step tutorials and the Python source code files for all examples.
Let’s get started.
- Updated Apr/2019: Updated the link to dataset.

How to Use Weight Regularization with LSTM Networks for Time Series Forecasting
Photo by Julian Fong, some rights reserved.
Tutorial Overview
This tutorial is broken down into 6 parts. They are:
- Shampoo Sales Dataset
- Experimental Test Harness
- Bias Weight Regularization
- Input Weight Regularization
- Recurrent Weight Regularization
- Review of Results
Environment
This tutorial assumes you have a Python SciPy environment installed. You can use either Python 2 or 3 with this example.
This tutorial assumes you have Keras v2.0 or higher installed with either the TensorFlow or Theano backend.
This tutorial also assumes you have scikit-learn, Pandas, NumPy, and Matplotlib installed.
If you need help setting up your Python environment, see this post:
Next, let’s take a look at a standard time series forecasting problem that we can use as context for this experiment.
Need help with Deep Learning for Time Series?
Take my free 7-day email crash course now (with sample code).
Click to sign-up and also get a free PDF Ebook version of the course.
Shampoo Sales Dataset
This dataset describes the monthly number of sales of shampoo over a 3-year period.
The units are a sales count and there are 36 observations. The original dataset is credited to Makridakis, Wheelwright, and Hyndman (1998).
The example below loads and creates a plot of the loaded dataset.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 |
# load and plot dataset from pandas import read_csv from pandas import datetime from matplotlib import pyplot # load dataset def parser(x): return datetime.strptime('190'+x, '%Y-%m') series = read_csv('shampoo-sales.csv', header=0, parse_dates=[0], index_col=0, squeeze=True, date_parser=parser) # summarize first few rows print(series.head()) # line plot series.plot() pyplot.show() |
Running the example loads the dataset as a Pandas Series and prints the first 5 rows.
|
1 2 3 4 5 6 7 |
Month 1901-01-01 266.0 1901-02-01 145.9 1901-03-01 183.1 1901-04-01 119.3 1901-05-01 180.3 Name: Sales, dtype: float64 |
A line plot of the series is then created showing a clear increasing trend.
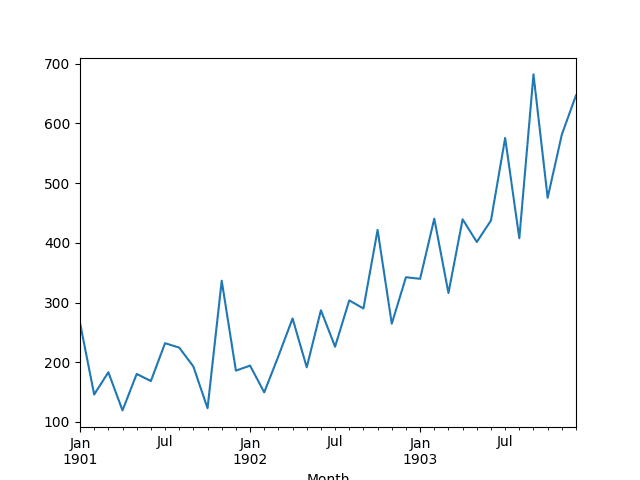
Line Plot of Shampoo Sales Dataset
Next, we will take a look at the model configuration and test harness used in the experiment.
Experimental Test Harness
This section describes the test harness used in this tutorial.
Data Split
We will split the Shampoo Sales dataset into two parts: a training and a test set.
The first two years of data will be taken for the training dataset and the remaining one year of data will be used for the test set.
Models will be developed using the training dataset and will make predictions on the test dataset.
The persistence forecast (naive forecast) on the test dataset achieves an error of 136.761 monthly shampoo sales. This provides a lower acceptable bound of performance on the test set.
Model Evaluation
A rolling-forecast scenario will be used, also called walk-forward model validation.
Each time step of the test dataset will be walked one at a time. A model will be used to make a forecast for the time step, then the actual expected value from the test set will be taken and made available to the model for the forecast on the next time step.
This mimics a real-world scenario where new Shampoo Sales observations would be available each month and used in the forecasting of the following month.
This will be simulated by the structure of the train and test datasets.
All forecasts on the test dataset will be collected and an error score calculated to summarize the skill of the model. The root mean squared error (RMSE) will be used as it punishes large errors and results in a score that is in the same units as the forecast data, namely monthly shampoo sales.
Data Preparation
Before we can fit a model to the dataset, we must transform the data.
The following three data transforms are performed on the dataset prior to fitting a model and making a forecast.
- Transform the time series data so that it is stationary. Specifically, a lag=1 differencing to remove the increasing trend in the data.
- Transform the time series into a supervised learning problem. Specifically, the organization of data into input and output patterns where the observation at the previous time step is used as an input to forecast the observation at the current timestep
- Transform the observations to have a specific scale. Specifically, to rescale the data to values between -1 and 1.
These transforms are inverted on forecasts to return them into their original scale before calculating and error score.
LSTM Model
We will use a base stateful LSTM model with 1 neuron fit for 1000 epochs.
Ideally a batch size of 1 would be used for walk-foward validation. We will assume walk-forward validation and predict the whole year for speed. As such we can use any batch size that is divisble by the number of samples, in this case we will use a value of 4.
Ideally, more training epochs would be used (such as 1500), but this was truncated to 1000 to keep run times reasonable.
The model will be fit using the efficient ADAM optimization algorithm and the mean squared error loss function.
Experimental Runs
Each experimental scenario will be run 30 times and the RMSE score on the test set will be recorded from the end each run.
Let’s dive into the experiments.
Baseline LSTM Model
Let’s start-off with the baseline LSTM model.
The baseline LSTM model for this problem has the following configuration:
- Lag inputs: 1
- Epochs: 1000
- Units in LSTM hidden layer: 3
- Batch Size: 4
- Repeats: 3
The complete code listing is provided below.
This code listing will be used as the basis for all following experiments, with only the changes to this code provided in subsequent sections.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 |
from pandas import DataFrame from pandas import Series from pandas import concat from pandas import read_csv from pandas import datetime from sklearn.metrics import mean_squared_error from sklearn.preprocessing import MinMaxScaler from keras.models import Sequential from keras.layers import Dense from keras.layers import LSTM from keras.regularizers import L1L2 from math import sqrt import matplotlib # be able to save images on server matplotlib.use('Agg') from matplotlib import pyplot import numpy # date-time parsing function for loading the dataset def parser(x): return datetime.strptime('190'+x, '%Y-%m') # frame a sequence as a supervised learning problem def timeseries_to_supervised(data, lag=1): df = DataFrame(data) columns = [df.shift(i) for i in range(1, lag+1)] columns.append(df) df = concat(columns, axis=1) return df # create a differenced series def difference(dataset, interval=1): diff = list() for i in range(interval, len(dataset)): value = dataset[i] - dataset[i - interval] diff.append(value) return Series(diff) # invert differenced value def inverse_difference(history, yhat, interval=1): return yhat + history[-interval] # scale train and test data to [-1, 1] def scale(train, test): # fit scaler scaler = MinMaxScaler(feature_range=(-1, 1)) scaler = scaler.fit(train) # transform train train = train.reshape(train.shape[0], train.shape[1]) train_scaled = scaler.transform(train) # transform test test = test.reshape(test.shape[0], test.shape[1]) test_scaled = scaler.transform(test) return scaler, train_scaled, test_scaled # inverse scaling for a forecasted value def invert_scale(scaler, X, yhat): new_row = [x for x in X] + [yhat] array = numpy.array(new_row) array = array.reshape(1, len(array)) inverted = scaler.inverse_transform(array) return inverted[0, -1] # fit an LSTM network to training data def fit_lstm(train, n_batch, nb_epoch, n_neurons): X, y = train[:, 0:-1], train[:, -1] X = X.reshape(X.shape[0], 1, X.shape[1]) model = Sequential() model.add(LSTM(n_neurons, batch_input_shape=(n_batch, X.shape[1], X.shape[2]), stateful=True)) model.add(Dense(1)) model.compile(loss='mean_squared_error', optimizer='adam') for i in range(nb_epoch): model.fit(X, y, epochs=1, batch_size=n_batch, verbose=0, shuffle=False) model.reset_states() return model # run a repeated experiment def experiment(series, n_lag, n_repeats, n_epochs, n_batch, n_neurons): # transform data to be stationary raw_values = series.values diff_values = difference(raw_values, 1) # transform data to be supervised learning supervised = timeseries_to_supervised(diff_values, n_lag) supervised_values = supervised.values[n_lag:,:] # split data into train and test-sets train, test = supervised_values[0:-12], supervised_values[-12:] # transform the scale of the data scaler, train_scaled, test_scaled = scale(train, test) # run experiment error_scores = list() for r in range(n_repeats): # fit the model train_trimmed = train_scaled[2:, :] lstm_model = fit_lstm(train_trimmed, n_batch, n_epochs, n_neurons) # forecast test dataset test_reshaped = test_scaled[:,0:-1] test_reshaped = test_reshaped.reshape(len(test_reshaped), 1, 1) output = lstm_model.predict(test_reshaped, batch_size=n_batch) predictions = list() for i in range(len(output)): yhat = output[i,0] X = test_scaled[i, 0:-1] # invert scaling yhat = invert_scale(scaler, X, yhat) # invert differencing yhat = inverse_difference(raw_values, yhat, len(test_scaled)+1-i) # store forecast predictions.append(yhat) # report performance rmse = sqrt(mean_squared_error(raw_values[-12:], predictions)) print('%d) Test RMSE: %.3f' % (r+1, rmse)) error_scores.append(rmse) return error_scores # configure the experiment def run(): # load dataset series = read_csv('shampoo-sales.csv', header=0, parse_dates=[0], index_col=0, squeeze=True, date_parser=parser) # configure the experiment n_lag = 1 n_repeats = 30 n_epochs = 1000 n_batch = 4 n_neurons = 3 # run the experiment results = DataFrame() results['results'] = experiment(series, n_lag, n_repeats, n_epochs, n_batch, n_neurons) # summarize results print(results.describe()) # save boxplot results.boxplot() pyplot.savefig('experiment_baseline.png') # entry point run() |
Running the experiment prints summary statistics for the test RMSE for all repeats.
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
We can see that on average, this model configuration achieves a test RMSE of about 92 monthly shampoo sales with a standard deviation of 5.
|
1 2 3 4 5 6 7 8 9 |
results count 30.000000 mean 92.842537 std 5.748456 min 81.205979 25% 89.514367 50% 92.030003 75% 96.926145 max 105.247117 |
A box and whisker plot is also created from the distribution of test RMSE results and saved to a file.
The plot provides a clear depiction of the spread of the results, highlighting the middle 50% of values (the box) and the median (green line).
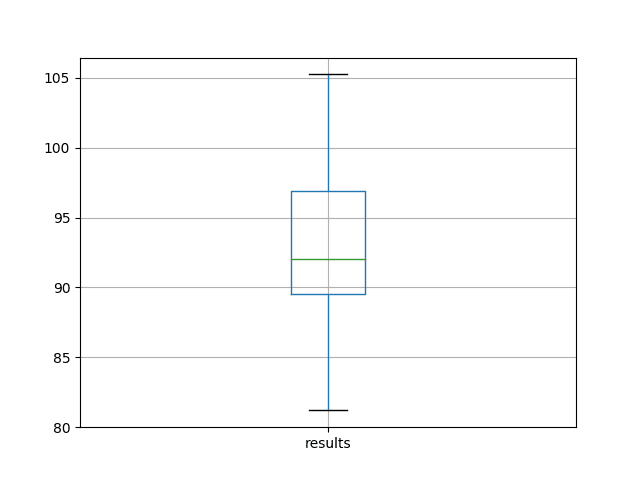
Box and Whisker Plot of Baseline Performance on the Shampoo Sales Dataset
Bias Weight Regularization
Weight regularization can be applied to the bias connection within the LSTM nodes.
In Keras, this is specified with a bias_regularizer argument when creating an LSTM layer. The regularizer is defined as an instance of the one of the L1, L2, or L1L2 classes.
More details here:
In this experiment, we will compare L1, L2, and L1L2 with a default value of 0.01 against the baseline model. We can specify all configurations using the L1L2 class, as follows:
- L1L2(0.0, 0.0) [e.g. baseline]
- L1L2(0.01, 0.0) [e.g. L1]
- L1L2(0.0, 0.01) [e.g. L2]
- L1L2(0.01, 0.01) [e.g. L1L2 or elasticnet]
Below lists the updated fit_lstm(), experiment(), and run() functions for using bias weight regularization with LSTMs.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 |
# fit an LSTM network to training data def fit_lstm(train, n_batch, nb_epoch, n_neurons, reg): X, y = train[:, 0:-1], train[:, -1] X = X.reshape(X.shape[0], 1, X.shape[1]) model = Sequential() model.add(LSTM(n_neurons, batch_input_shape=(n_batch, X.shape[1], X.shape[2]), stateful=True, bias_regularizer=reg)) model.add(Dense(1)) model.compile(loss='mean_squared_error', optimizer='adam') for i in range(nb_epoch): model.fit(X, y, epochs=1, batch_size=n_batch, verbose=0, shuffle=False) model.reset_states() return model # run a repeated experiment def experiment(series, n_lag, n_repeats, n_epochs, n_batch, n_neurons, reg): # transform data to be stationary raw_values = series.values diff_values = difference(raw_values, 1) # transform data to be supervised learning supervised = timeseries_to_supervised(diff_values, n_lag) supervised_values = supervised.values[n_lag:,:] # split data into train and test-sets train, test = supervised_values[0:-12], supervised_values[-12:] # transform the scale of the data scaler, train_scaled, test_scaled = scale(train, test) # run experiment error_scores = list() for r in range(n_repeats): # fit the model train_trimmed = train_scaled[2:, :] lstm_model = fit_lstm(train_trimmed, n_batch, n_epochs, n_neurons, reg) # forecast test dataset test_reshaped = test_scaled[:,0:-1] test_reshaped = test_reshaped.reshape(len(test_reshaped), 1, 1) output = lstm_model.predict(test_reshaped, batch_size=n_batch) predictions = list() for i in range(len(output)): yhat = output[i,0] X = test_scaled[i, 0:-1] # invert scaling yhat = invert_scale(scaler, X, yhat) # invert differencing yhat = inverse_difference(raw_values, yhat, len(test_scaled)+1-i) # store forecast predictions.append(yhat) # report performance rmse = sqrt(mean_squared_error(raw_values[-12:], predictions)) print('%d) Test RMSE: %.3f' % (r+1, rmse)) error_scores.append(rmse) return error_scores # configure the experiment def run(): # load dataset series = read_csv('shampoo-sales.csv', header=0, parse_dates=[0], index_col=0, squeeze=True, date_parser=parser) # configure the experiment n_lag = 1 n_repeats = 30 n_epochs = 1000 n_batch = 4 n_neurons = 3 regularizers = [L1L2(l1=0.0, l2=0.0), L1L2(l1=0.01, l2=0.0), L1L2(l1=0.0, l2=0.01), L1L2(l1=0.01, l2=0.01)] # run the experiment results = DataFrame() for reg in regularizers: name = ('l1 %.2f,l2 %.2f' % (reg.l1, reg.l2)) results[name] = experiment(series, n_lag, n_repeats, n_epochs, n_batch, n_neurons, reg) # summarize results print(results.describe()) # save boxplot results.boxplot() pyplot.savefig('experiment_reg_bias.png') |
Running this experiment prints descriptive statistics for each evaluated configuration.
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
The results suggest that on average, the default of no bias regularization results in better performance compared to all of the other configurations considered.
|
1 2 3 4 5 6 7 8 9 |
l1 0.00,l2 0.00 l1 0.01,l2 0.00 l1 0.00,l2 0.01 l1 0.01,l2 0.01 count 30.000000 30.000000 30.000000 30.000000 mean 92.821489 95.520003 93.285389 92.901021 std 4.894166 3.637022 3.906112 5.082358 min 81.394504 89.477398 82.994480 78.729224 25% 89.356330 93.017723 90.907343 91.210105 50% 92.822871 95.502700 93.837562 94.525965 75% 95.899939 98.195980 95.426270 96.882378 max 101.194678 101.750900 100.650130 97.766301 |
A box and whisker plot is also created to compare the distributions of results for each configuration.
The plot shows that all configurations have about the same spread and that the addition of bias regularization uniformly was not helpful on this problem.
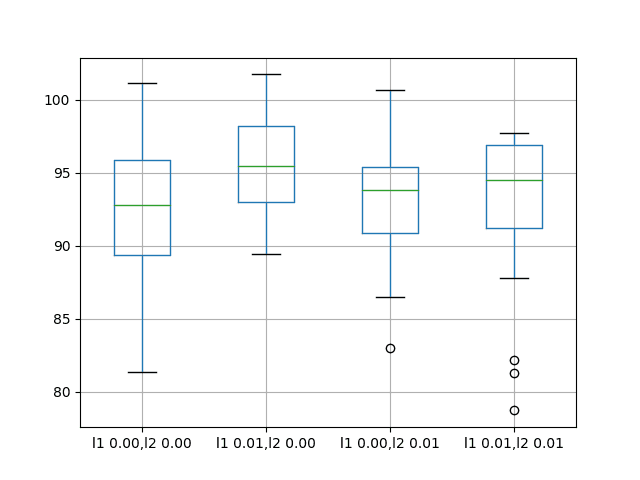
Box and Whisker Plots of Bias Weight Regularization Performance on the Shampoo Sales Dataset
Input Weight Regularization
We can also apply regularization to input connections on each LSTM unit.
In Keras, this is achieved by setting the kernel_regularizer argument to a regularizer class.
We will test the same regularizer configurations as were used in the previous section, specifically:
- L1L2(0.0, 0.0) [e.g. baseline]
- L1L2(0.01, 0.0) [e.g. L1]
- L1L2(0.0, 0.01) [e.g. L2]
- L1L2(0.01, 0.01) [e.g. L1L2 or elasticnet]
Below lists the updated fit_lstm(), experiment(), and run() functions for using bias weight regularization with LSTMs.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 |
# fit an LSTM network to training data def fit_lstm(train, n_batch, nb_epoch, n_neurons, reg): X, y = train[:, 0:-1], train[:, -1] X = X.reshape(X.shape[0], 1, X.shape[1]) model = Sequential() model.add(LSTM(n_neurons, batch_input_shape=(n_batch, X.shape[1], X.shape[2]), stateful=True, kernel_regularizer=reg)) model.add(Dense(1)) model.compile(loss='mean_squared_error', optimizer='adam') for i in range(nb_epoch): model.fit(X, y, epochs=1, batch_size=n_batch, verbose=0, shuffle=False) model.reset_states() return model # run a repeated experiment def experiment(series, n_lag, n_repeats, n_epochs, n_batch, n_neurons, reg): # transform data to be stationary raw_values = series.values diff_values = difference(raw_values, 1) # transform data to be supervised learning supervised = timeseries_to_supervised(diff_values, n_lag) supervised_values = supervised.values[n_lag:,:] # split data into train and test-sets train, test = supervised_values[0:-12], supervised_values[-12:] # transform the scale of the data scaler, train_scaled, test_scaled = scale(train, test) # run experiment error_scores = list() for r in range(n_repeats): # fit the model train_trimmed = train_scaled[2:, :] lstm_model = fit_lstm(train_trimmed, n_batch, n_epochs, n_neurons, reg) # forecast test dataset test_reshaped = test_scaled[:,0:-1] test_reshaped = test_reshaped.reshape(len(test_reshaped), 1, 1) output = lstm_model.predict(test_reshaped, batch_size=n_batch) predictions = list() for i in range(len(output)): yhat = output[i,0] X = test_scaled[i, 0:-1] # invert scaling yhat = invert_scale(scaler, X, yhat) # invert differencing yhat = inverse_difference(raw_values, yhat, len(test_scaled)+1-i) # store forecast predictions.append(yhat) # report performance rmse = sqrt(mean_squared_error(raw_values[-12:], predictions)) print('%d) Test RMSE: %.3f' % (r+1, rmse)) error_scores.append(rmse) return error_scores # configure the experiment def run(): # load dataset series = read_csv('shampoo-sales.csv', header=0, parse_dates=[0], index_col=0, squeeze=True, date_parser=parser) # configure the experiment n_lag = 1 n_repeats = 30 n_epochs = 1000 n_batch = 4 n_neurons = 3 regularizers = [L1L2(l1=0.0, l2=0.0), L1L2(l1=0.01, l2=0.0), L1L2(l1=0.0, l2=0.01), L1L2(l1=0.01, l2=0.01)] # run the experiment results = DataFrame() for reg in regularizers: name = ('l1 %.2f,l2 %.2f' % (reg.l1, reg.l2)) results[name] = experiment(series, n_lag, n_repeats, n_epochs, n_batch, n_neurons, reg) # summarize results print(results.describe()) # save boxplot results.boxplot() pyplot.savefig('experiment_reg_input.png') |
Running this experiment prints descriptive statistics for each evaluated configuration.
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
The results suggest that adding weight regularization to input connections does offer benefit across the board on this setup.
We can see that the test RMSE is approximately 10 units lower for all configurations with perhaps more benefit when both L1 and L2 are combined into an elasticnet type constraint.
|
1 2 3 4 5 6 7 8 9 |
l1 0.00,l2 0.00 l1 0.01,l2 0.00 l1 0.00,l2 0.01 l1 0.01,l2 0.01 count 30.000000 30.000000 30.000000 30.000000 mean 91.640028 82.118980 82.137198 80.471685 std 6.086401 4.116072 3.378984 2.212213 min 80.392310 75.705210 76.005173 76.550909 25% 88.025135 79.237822 79.698162 78.780802 50% 91.843761 81.235433 81.463882 80.283913 75% 94.860117 85.510177 84.563980 82.443390 max 105.820586 94.210503 90.823454 85.243135 |
A box and whisker plot is also created to compare the distributions of results for each configuration.
The plot shows the general lower distribution of error for input regularization. The results also suggest a tighter spread of results with regularization that may be more pronounced with the L1L2 configuration that achieved the better results.
This is an encouraging finding, suggesting that additional experiments with different L1L2 values for input regularization would be well worth investigating.
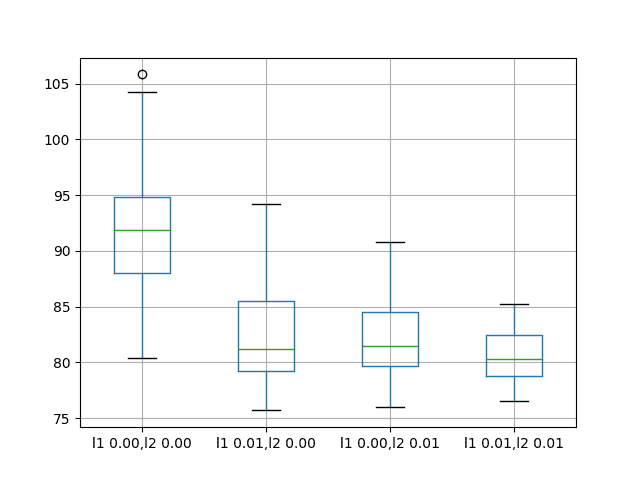
Box and Whisker Plots of Input Weight Regularization Performance on the Shampoo Sales Dataset
Recurrent Weight Regularization
Finally, we can also apply regularization to recurrent connections on each LSTM unit.
In Keras, this is achieved by setting the recurrent_regularizer argument to a regularizer class.
We will test the same regularizer configurations as were used in the previous section, specifically:
- L1L2(0.0, 0.0) [e.g. baseline]
- L1L2(0.01, 0.0) [e.g. L1]
- L1L2(0.0, 0.01) [e.g. L2]
- L1L2(0.01, 0.01) [e.g. L1L2 or elasticnet]
Below lists the updated fit_lstm(), experiment(), and run() functions for using bias weight regularization with LSTMs.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 |
# fit an LSTM network to training data def fit_lstm(train, n_batch, nb_epoch, n_neurons, reg): X, y = train[:, 0:-1], train[:, -1] X = X.reshape(X.shape[0], 1, X.shape[1]) model = Sequential() model.add(LSTM(n_neurons, batch_input_shape=(n_batch, X.shape[1], X.shape[2]), stateful=True, recurrent_regularizer=reg)) model.add(Dense(1)) model.compile(loss='mean_squared_error', optimizer='adam') for i in range(nb_epoch): model.fit(X, y, epochs=1, batch_size=n_batch, verbose=0, shuffle=False) model.reset_states() return model # run a repeated experiment def experiment(series, n_lag, n_repeats, n_epochs, n_batch, n_neurons, reg): # transform data to be stationary raw_values = series.values diff_values = difference(raw_values, 1) # transform data to be supervised learning supervised = timeseries_to_supervised(diff_values, n_lag) supervised_values = supervised.values[n_lag:,:] # split data into train and test-sets train, test = supervised_values[0:-12], supervised_values[-12:] # transform the scale of the data scaler, train_scaled, test_scaled = scale(train, test) # run experiment error_scores = list() for r in range(n_repeats): # fit the model train_trimmed = train_scaled[2:, :] lstm_model = fit_lstm(train_trimmed, n_batch, n_epochs, n_neurons, reg) # forecast test dataset test_reshaped = test_scaled[:,0:-1] test_reshaped = test_reshaped.reshape(len(test_reshaped), 1, 1) output = lstm_model.predict(test_reshaped, batch_size=n_batch) predictions = list() for i in range(len(output)): yhat = output[i,0] X = test_scaled[i, 0:-1] # invert scaling yhat = invert_scale(scaler, X, yhat) # invert differencing yhat = inverse_difference(raw_values, yhat, len(test_scaled)+1-i) # store forecast predictions.append(yhat) # report performance rmse = sqrt(mean_squared_error(raw_values[-12:], predictions)) print('%d) Test RMSE: %.3f' % (r+1, rmse)) error_scores.append(rmse) return error_scores # configure the experiment def run(): # load dataset series = read_csv('shampoo-sales.csv', header=0, parse_dates=[0], index_col=0, squeeze=True, date_parser=parser) # configure the experiment n_lag = 1 n_repeats = 30 n_epochs = 1000 n_batch = 4 n_neurons = 3 regularizers = [L1L2(l1=0.0, l2=0.0), L1L2(l1=0.01, l2=0.0), L1L2(l1=0.0, l2=0.01), L1L2(l1=0.01, l2=0.01)] # run the experiment results = DataFrame() for reg in regularizers: name = ('l1 %.2f,l2 %.2f' % (reg.l1, reg.l2)) results[name] = experiment(series, n_lag, n_repeats, n_epochs, n_batch, n_neurons, reg) # summarize results print(results.describe()) # save boxplot results.boxplot() pyplot.savefig('experiment_reg_recurrent.png') |
Running this experiment prints descriptive statistics for each evaluated configuration.
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
The results suggest no obvious benefit from using regularization on the recurrent connection with LSTMs on this problem.
The mean performance of all variations tried resulted in worse performance than the baseline model.
|
1 2 3 4 5 6 7 8 9 |
l1 0.00,l2 0.00 l1 0.01,l2 0.00 l1 0.00,l2 0.01 l1 0.01,l2 0.01 count 30.000000 30.000000 30.000000 30.000000 mean 92.918797 100.675386 101.302169 96.820026 std 7.172764 3.866547 5.228815 3.966710 min 72.967841 93.789854 91.063592 89.367600 25% 90.311185 98.014045 98.222732 94.787647 50% 92.327824 100.546756 101.903350 95.727549 75% 95.989761 103.491192 104.918266 98.240613 max 110.529422 108.788604 110.712064 111.185747 |
A box and whisker plot is also created to compare the distributions of results for each configuration.
The plot shows the same story as the summary statistics, suggesting little benefit from using recurrent weight regularization.
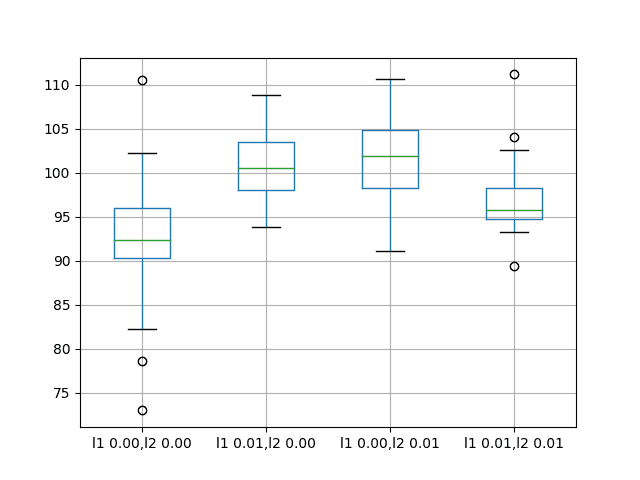
Box and Whisker Plots of Recurrent Weight Regularization Performance on the Shampoo Sales Dataset
Extensions
This section lists ideas for follow-up experiments to extend the work in this tutorial.
- Input Weight Regularization. The experimental results for input weight regularization on this problem showed great promise of listing performance. This could be investigated further by perhaps grid searching different L1 and L2 values to find an optimal configuration.
- Behavior Dynamics. The dynamic behavior of each weight regularization scheme could be investigated by plotting train and test RMSE over training epochs to get an idea of weight regularization on overfitting or underfitting behavior patterns.
- Combine Regularization. Experiments could be designed to explore the effect of combining different weight regularization schemes.
- Activation Regularization. Keras also supports activation regularization, and this could be another avenue to explore imposing constraints on the LSTM and reduce overfitting.
Summary
In this tutorial, you discovered how to use weight regularization with LSTMs for time series forecasting.
Specifically, you learned:
- How to design a robust test harness for evaluating LSTM networks for time series forecasting.
- How to configure bias weight regularization on LSTMs for time series forecasting.
- How to configure input and recurrent weight regularization on LSTMs for time series forecasting.
Do you have any questions about using weight regularization with LSTM networks?
Ask your questions in the comments below and I will do my best to answer.








Gret job like every time.
But I want to understand why always we are using rmse and we don’t use the accuracy metrics ??
You cannot measure accuracy on regression problems (unless you transform them to become a classification problem).
In general, problems that have a real-valued quantity as the output variable are regression problems, those that have a category or label output are classification.
Ah OK, yes it is a logic thinking. Thanks
hello jason,
how can be predicted a stock market data using a multiple input of columns and single output columns.
Each separate time series would be framed as a separate feature in the LSTM.
also, I believe short term security prices are a random walk and are not predictable:
https://machinelearningmastery.com/gentle-introduction-random-walk-times-series-forecasting-python/
import numpy as np
import pandas as pd
import tensorflow as tf
#tf.logging.set_verosity(tf.logging.ERROR)
from pandas_datareader import data as web
import matplotlib.pyplot as plt
def get_data():
feature_cols={‘ret_%s’ %i:tf.constant(idata[‘ret_%s’%i].values)for i in lags}
labels=tf.constant((idata[‘returns’]>0).astype(int).values,shape=[idata[‘returns’].size,1])
return feature_cols,labels
symbol=’^GSPC’
data=web.DataReader(symbol,data_source=’yahoo’,start=’2014-01-01′,end=’2016-10-31′)[‘Adj Close’]
data=pd.DataFrame(data)
data.rename(columns={‘Adj Close’:’price’},inplace=True)
data[‘returns’]=np.log(data/data.shift(1))
lags=range(1,6)
for i in lags:
data[‘ret_%s’% i]=np.sign(data[‘returns’].shift(i))
data.dropna(inplace=True)
print data.round(4).tail()
cutoff=’2015-1-1′
training_data=data[data.index=cutoff].copy()
#def get_data():
#feature_cols={‘ret_%s’ %i:tf.constant(data[‘ret_%s’%i].values)for i in lags}
#labels=tf.constant((data[‘returns’]>0).astype(int).values,shape=[data[‘returns’].size,1])
#return feature_cols,labels
fc=[tf.contrib.layers.real_valued_column(‘ret_%s’% i,dimension=1) for i in lags]
model=tf.contrib.learn.DNNClassifier(feature_columns=fc,n_classes=2,hidden_units=[100,100])
idata=training_data
model.fit(input_fn=get_data,steps=500)
model.evaluate(input_fn=get_data,steps=1)
pred=model.predict(input_fn=get_data)
pred[:30]
training_data[‘prediction’]=np.where(pred>0,1,-1)
training_data[‘strategy’]=training_data[‘prediction’]*training_data[‘returns’]
training_data[[‘returns’,’strategy’]].cumsum().apply(np.exp).plot(figsize=(10,6))
idata=test_data
model.evaluate(input_fn=get_data,steps=1)
pred=model.predict(input_fn=get_data)
test_data[‘prediction’]=np.where(pred>0,1,-1)
test_data[‘strategy’]=test_data[‘prediction’]*test_data[‘returns’]
test_data[[‘returns’,’strategy’]].cumsum().apply(np.exp).plot(figsize=(10,6))
#pred[:1]
if __name__ == ‘__main__’:
get_data()
hello jason,
i just ran this code but i got some error in the code and i cannot understand the error.
the error is:—–> 1 pred[:30]
TypeError: ‘generator’ object is not subscriptable
please kindly help me.
Perhaps contact the author of the tensorflow code you have pasted?
Great article!
however, what confused me is, in section LSTM model, “A batch size of 1 is required as we will be using walk-forward validation and making one-step forecasts for each of the final 12 months of test data.”, but then you use batch size as 4 (n_batch=4).
I guess you mean “A time step of 1 is required”, am I right? cause one forecast per month means one time step.
You make a good point.
A batch size of 1 would be required if we made predictions each time step of the test data. Here we predict the whole year first, then work through the predictions to see what they mean.
I have updated the post correcting the error.
Thanks for all your pedagogic material, it’s really easy to learn with your articles!
I have a question: in all your examples of time series deep learning, you’re always predicting the next time step, one at a time.
How would you approach the case of training with a time series, and forecasting the next 12 steps, for example?
Here is an example:
https://machinelearningmastery.com/multi-step-time-series-forecasting-long-short-term-memory-networks-python/
HI, your article is very useful for me! But I run the “Bias Weight Regularization procedure”, the spyder will have the fault :”D:\Program Files\Anaconda3\lib\site-packages\matplotlib\__init__.py:1357: UserWarning: This call to matplotlib.use() has no effect
because the backend has already been chosen;
matplotlib.use() must be called *before* pylab, matplotlib.pyplot,
or matplotlib.backends is imported for the first time.
warnings.warn(_use_error_msg)”,
I don’t know how to slove the fault, could you help me ?
Are you able to run the script from the command line instead of within the IDE?
Thank you very much! Because I missed the command
“# entry point
run()”.
When I added the command,the procedure could run.
I’m glad to hear it.
I have one doubt related to stateful = True. As mentioned in keras documentation, first element of first batch is in sequence with first element of second batch and so on. But you have not converted time series into that order. Is it correct observation or I am missing something?
I’m pretty sure the implementation is wrong. If you check on the keras manual, the input to the LSTM should be (batch_size, timesteps, input_dim). The thing is that in this example we only have one sequence, so batch_size should be 1 and timesteps should be 4, not the opposite as shown in the article. Then it also makes sense that the state is kept between batches.
I’m trying to understand how you’re attaching the regularizers to the model. In the fit_lstm(…) methods you’re never actually attaching anything in the reg variable to the model. Or, am I missing something? Thank you!
They are specified on the LSTM layer via arguments.
Thank you! The code view widget was hiding it, my bad.
Seth
Thanks for the post. Your posts are extremely valuable and I appreciate the time you are taking to write these.
Why did you choose the number of hidden layers in a LSTM as just 3? In general how does one decide to choose the number of hidden units?
Great question. I used a little trial and error.
For more on configuring neural nets, see this:
https://machinelearningmastery.com/faq/single-faq/how-many-layers-and-nodes-do-i-need-in-my-neural-network
Thanks.
I am wondering if the number of hidden layers in LSTM is a function of the number of input samples to the model? ie each hidden layer has weights that needs to be learnt and if there is less data then there would be a tendency to overfit if we choose a large number of hidden layers.
On another note .. What is the dimension of the hidden layer ? For instance if the the input shape is (5,30) — (num of time steps, num of features) and the number of samples is 100,000 .. what would be the dimension of the first hidden layer and subsequent layers in LSTM
Regards,
-Avi
No. Units in the first hidden layer has nothing to do with the length of input sequences.
You can see the shape of each layer by printing the output of model.summary().
Hi Jason,
Thanks for sharing this article.
I have an LSTM network having 3 LSTM layers. I was wondering if I need to add recurrent regularization at each LSTM layer?
Try it and see.
I’ve seen the good contents.
May I ask you a question personally? Can you emphasize specific variables in your predictions with LSTM?
When model calculate the weight of a variable, can you make a particular variable more important?
Thanks.
No, the model will figure out what is most important during learning.
Hey thanks for nice explaination!!
My question is
Is there any difference in your bias weight regularization code and input weight regularization code because in both the way of use regularization is similar, how will the system identify that you want to use bias regularization or input weight regularization??
Like for recurrent_regularizer you have specified it but what about input weight regularization
ohhh!! I got the answer
Thanks man for the nice blog
No problem.
Different arguments are used on the LSTM layers for bias vs weight regularization.
Hello, Dr. Brownlee,
I have a question on validation:
What would be the best way to incorporate validation_split or validation_data parameter into the model.fit() with the setup you have outlined above? I.e. perform validation during training to asses over-fit.
validation_split does not seem to work…
Thanks
Validation data for LSTMs is very tricky. You might be better off using walk-forward validation.
hI,
Do you have any article about the inner procedures when we apply bias/weight or recurrent regularization? Thanks in advanced.
in other words which component is best to regularize?
I recommend comparing the results from a suite of regularization techniques.
This might help as a starting point:
https://machinelearningmastery.com/introduction-to-regularization-to-reduce-overfitting-and-improve-generalization-error/
Extensions
This section lists ideas for follow-up experiments to extend the work in this tutorial.
Activation Regularization. Keras also supports activation regularization, and this could be another avenue to explore imposing constraints on the LSTM and reduce overfitting.
hi jason,
I read above work .in that you gave some extensions ….In particluar last work is activation regularization (activation function regularization ) is that correct . which means can i use regualrization techniques in the activation function in order to handle the overfitting concepts…way which i understood correct or wrong ..
Yes, here is an example:
https://machinelearningmastery.com/how-to-reduce-generalization-error-in-deep-neural-networks-with-activity-regularization-in-keras/
hi jason,
Can i use both weight regularization in LSTM and activation regularization in the HIDDEN LAYER at the same time in order to overcome the overfitting…
I don’t see why not, perhaps try it and compare results to not using it.
hi jason,
is that possible to use lstm+ weight regularization +dropout techniques together to reduce overfitting….
Yes, perhaps try the combination to see if it performs better on your dataset.
hi jason,
is that possible to use activation regularization on LSTM Nodes in order to overcome overfitting
It can be used with LSTMs.
It may or may not help with overfitting depending on the specifics of your model and dataset.
hi jason ,
which regularization techniques L1(OR) L2 (OR) ELASTIC NET IS MORE SUITABLE ONE ON LSTM MODEL..
It depends on the specifics of your model and dataset.
Perhaps try a suite of approaches and discover what works best for you.
hi jason,
using L1 regularization on lstm output layer and dropout in hidden (dense) layer to build a generalised model
That sounds odd. Typically only one regularization method would be used in a model.
Hi Jason,
If you find that your deep model needs Batch Normalization, but still needs some regularization to prevent over fitting. What methods would apply to test?
I have read that Dropouts shouldn’t be used together with Batch Norm, also I have read that (at least some) methods of Lx regulation takes another role when combined with Batch Norm (https://blog.janestreet.com/l2-regularization-and-batch-norm/).
From your experience, what should I start experimenting with in a deep LSTM network with Batch Normalization, where I still experience over fitting?
Lets also assume that getting more data to train on is exhausted, and augmenting data is a very limited option.
Dropout still works well, so does early stopping and weight decay.
Also slow down learning with a smaller learning rate and more epochs.
Generally, test a few methods and discover what works well/best for your model + data.
hi jason,
does Lstm need feature extraction and feature selection of EEG signals… or automatically learn the required features from EEG signals
It can learn to extract features from the signal.
hi jason,
deep learning model (LSTM ) requires any explicit feature selection and feature extraction techniques when using EEG dataset
It may or may not. Try with and without and compare the performance of the model on your dataset. Use what works best.
Dear Prof. Jason Brownlee, thank you for this tutorial and explanation.
I want to ask, can we use this Weight Regularization-LSTM to classify something? for example, for HAR as also found on this website.
If so, are the steps the same?
I’m still just learning about LSTM and python, so sorry if my question is too basic.
Thank you very much.
Should work but LSTM is designed for time series so the network can remember something. Is that useful for classification? You can try out to confirm.
Great article!!!.
Could you explain me why you use: X.reshape(X.shape[0], 1, X.shape[1])?
Thank you
Hi Jose…Please see the following that relates to reshaping input data:
https://machinelearningmastery.com/reshape-input-data-long-short-term-memory-networks-keras/
hey!! can we modify the weight of LSTM using a function?
weight=f(x,y)
then, assigned it to model
Hi Akkis…The following resource may be of interest:
https://machinelearningmastery.com/update-lstm-networks-training-time-series-forecasting/
How can one deduce which weights (and related to which inputs) have been regularized?
Hi Andrea…The following resource may be of interest to you:
https://machinelearningmastery.com/weight-regularization-to-reduce-overfitting-of-deep-learning-models/