Model averaging is an ensemble technique where multiple sub-models contribute equally to a combined prediction.
Model averaging can be improved by weighting the contributions of each sub-model to the combined prediction by the expected performance of the submodel. This can be extended further by training an entirely new model to learn how to best combine the contributions from each submodel. This approach is called stacked generalization, or stacking for short, and can result in better predictive performance than any single contributing model.
In this tutorial, you will discover how to develop a stacked generalization ensemble for deep learning neural networks.
After completing this tutorial, you will know:
- Stacked generalization is an ensemble method where a new model learns how to best combine the predictions from multiple existing models.
- How to develop a stacking model using neural networks as a submodel and a scikit-learn classifier as the meta-learner.
- How to develop a stacking model where neural network sub-models are embedded in a larger stacking ensemble model for training and prediction.
Kick-start your project with my new book Better Deep Learning, including step-by-step tutorials and the Python source code files for all examples.
Let’s get started.
- Updated Oct/2019: Updated for Keras 2.3 and TensorFlow 2.0.
- Update Jan/2020: Updated for changes in scikit-learn v0.22 API.
- Update Aug/2020: Updated for Keras 2.4.3 and TensorFlow 2.3

How to Develop a Stacking Ensemble for Deep Learning Neural Networks in Python With Keras
Photo by David Law, some rights reserved.
Tutorial Overview
This tutorial is divided into six parts; they are:
- Stacked Generalization Ensemble
- Multi-Class Classification Problem
- Multilayer Perceptron Model
- Train and Save Sub-Models
- Separate Stacking Model
- Integrated Stacking Model
Stacked Generalization Ensemble
A model averaging ensemble combines the predictions from multiple trained models.
A limitation of this approach is that each model contributes the same amount to the ensemble prediction, regardless of how well the model performed. A variation of this approach, called a weighted average ensemble, weighs the contribution of each ensemble member by the trust or expected performance of the model on a holdout dataset. This allows well-performing models to contribute more and less-well-performing models to contribute less. The weighted average ensemble provides an improvement over the model average ensemble.
A further generalization of this approach is replacing the linear weighted sum (e.g. linear regression) model used to combine the predictions of the sub-models with any learning algorithm. This approach is called stacked generalization, or stacking for short.
In stacking, an algorithm takes the outputs of sub-models as input and attempts to learn how to best combine the input predictions to make a better output prediction.
It may be helpful to think of the stacking procedure as having two levels: level 0 and level 1.
- Level 0: The level 0 data is the training dataset inputs and level 0 models learn to make predictions from this data.
- Level 1: The level 1 data takes the output of the level 0 models as input and the single level 1 model, or meta-learner, learns to make predictions from this data.
Stacked generalization works by deducing the biases of the generalizer(s) with respect to a provided learning set. This deduction proceeds by generalizing in a second space whose inputs are (for example) the guesses of the original generalizers when taught with part of the learning set and trying to guess the rest of it, and whose output is (for example) the correct guess.
— Stacked generalization, 1992.
Unlike a weighted average ensemble, a stacked generalization ensemble can use the set of predictions as a context and conditionally decide to weigh the input predictions differently, potentially resulting in better performance.
Interestingly, although stacking is described as an ensemble learning method with two or more level 0 models, it can be used in the case where there is only a single level 0 model. In this case, the level 1, or meta-learner, model learns to correct the predictions from the level 0 model.
… although it can also be used when one has only a single generalizer, as a technique to improve that single generalizer
— Stacked generalization, 1992.
It is important that the meta-learner is trained on a separate dataset to the examples used to train the level 0 models to avoid overfitting.
A simple way that this can be achieved is by splitting the training dataset into a train and validation set. The level 0 models are then trained on the train set. The level 1 model is then trained using the validation set, where the raw inputs are first fed through the level 0 models to get predictions that are used as inputs to the level 1 model.
A limitation of the hold-out validation set approach to training a stacking model is that level 0 and level 1 models are not trained on the full dataset.
A more sophisticated approach to training a stacked model involves using k-fold cross-validation to develop the training dataset for the meta-learner model. Each level 0 model is trained using k-fold cross-validation (or even leave-one-out cross-validation for maximum effect); the models are then discarded, but the predictions are retained. This means for each model, there are predictions made by a version of the model that was not trained on those examples, e.g. like having holdout examples, but in this case for the entire training dataset.
The predictions are then used as inputs to train the meta-learner. Level 0 models are then trained on the entire training dataset and together with the meta-learner, the stacked model can be used to make predictions on new data.
In practice, it is common to use different algorithms to prepare each of the level 0 models, to provide a diverse set of predictions.
… stacking is not normally used to combine models of the same type […] it is applied to models built by different learning algorithms.
— Practical Machine Learning Tools and Techniques, Second Edition, 2005.
It is also common to use a simple linear model to combine the predictions. Because use of a linear model is common, stacking is more recently referred to as “model blending” or simply “blending,” especially in machine learning competitions.
… the multi-response least squares linear regression technique should be employed as the high-level generalizer. This technique provides a method of combining level-0 models’ confidence
— Issues in Stacked Generalization, 1999.
A stacked generalization ensemble can be developed for regression and classification problems. In the case of classification problems, better results have been seen when using the prediction of class probabilities as input to the meta-learner instead of class labels.
… class probabilities should be used instead of the single predicted class as input attributes for higher-level learning. The class probabilities serve as the confidence measure for the prediction made.
— Issues in Stacked Generalization, 1999.
Now that we are familiar with stacked generalization, we can work through a case study of developing a stacked deep learning model.
Want Better Results with Deep Learning?
Take my free 7-day email crash course now (with sample code).
Click to sign-up and also get a free PDF Ebook version of the course.
Multi-Class Classification Problem
We will use a small multi-class classification problem as the basis to demonstrate the stacking ensemble.
The scikit-learn class provides the make_blobs() function that can be used to create a multi-class classification problem with the prescribed number of samples, input variables, classes, and variance of samples within a class.
The problem has two input variables (to represent the x and y coordinates of the points) and a standard deviation of 2.0 for points within each group. We will use the same random state (seed for the pseudorandom number generator) to ensure that we always get the same data points.
|
1 2 |
# generate 2d classification dataset X, y = make_blobs(n_samples=1000, centers=3, n_features=2, cluster_std=2, random_state=2) |
The results are the input and output elements of a dataset that we can model.
In order to get a feeling for the complexity of the problem, we can graph each point on a two-dimensional scatter plot and color each point by class value.
The complete example is listed below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 |
# scatter plot of blobs dataset from sklearn.datasets import make_blobs from matplotlib import pyplot from pandas import DataFrame # generate 2d classification dataset X, y = make_blobs(n_samples=1000, centers=3, n_features=2, cluster_std=2, random_state=2) # scatter plot, dots colored by class value df = DataFrame(dict(x=X[:,0], y=X[:,1], label=y)) colors = {0:'red', 1:'blue', 2:'green'} fig, ax = pyplot.subplots() grouped = df.groupby('label') for key, group in grouped: group.plot(ax=ax, kind='scatter', x='x', y='y', label=key, color=colors[key]) pyplot.show() |
Running the example creates a scatter plot of the entire dataset. We can see that the standard deviation of 2.0 means that the classes are not linearly separable (separable by a line) causing many ambiguous points.
This is desirable as it means that the problem is non-trivial and will allow a neural network model to find many different “good enough” candidate solutions, resulting in a high variance.
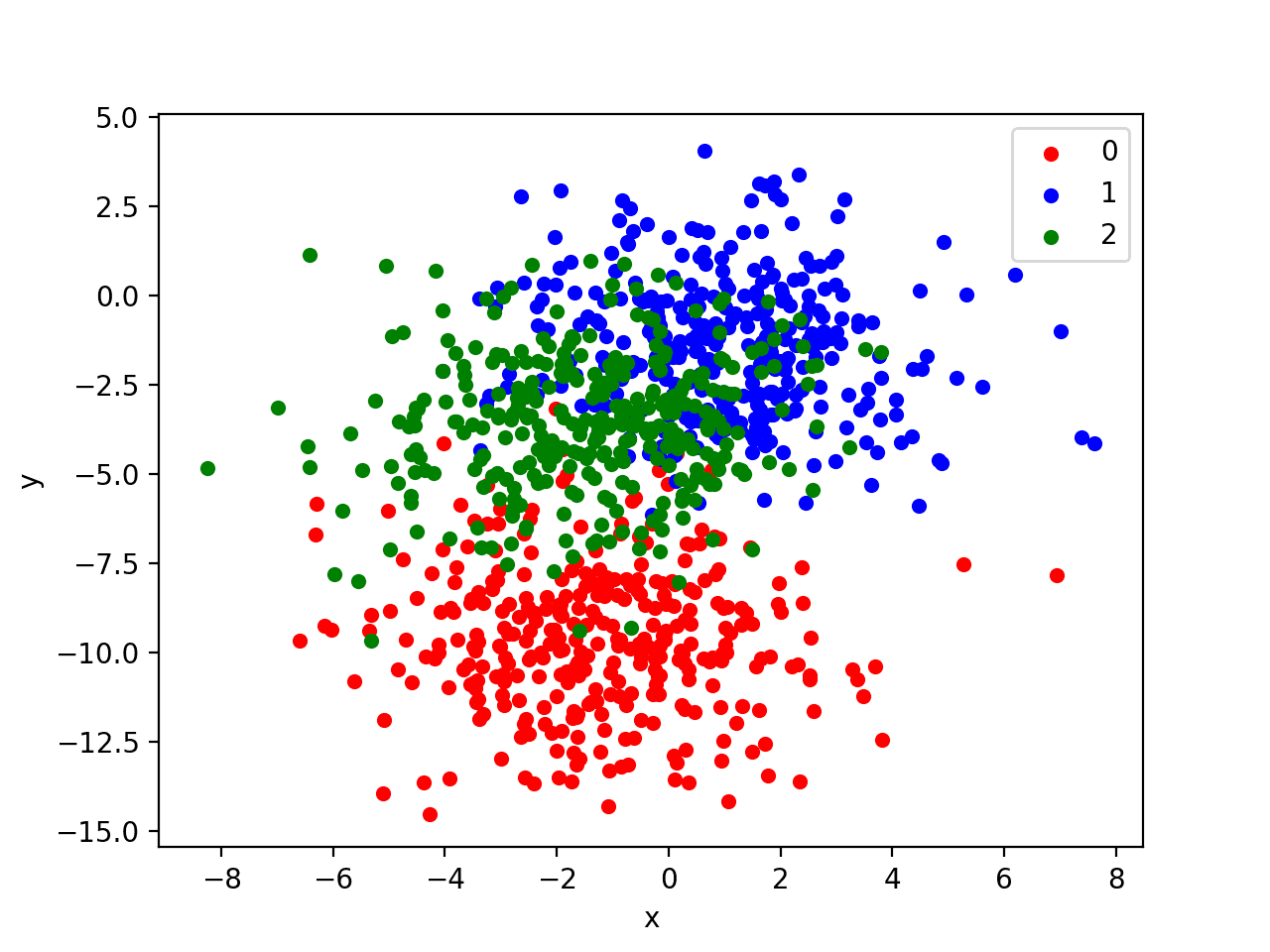
Scatter Plot of Blobs Dataset With Three Classes and Points Colored by Class Value
Multilayer Perceptron Model
Before we define a model, we need to contrive a problem that is appropriate for the stacking ensemble.
In our problem, the training dataset is relatively small. Specifically, there is a 10:1 ratio of examples in the training dataset to the holdout dataset. This mimics a situation where we may have a vast number of unlabeled examples and a small number of labeled examples with which to train a model.
We will create 1,100 data points from the blobs problem. The model will be trained on the first 100 points and the remaining 1,000 will be held back in a test dataset, unavailable to the model.
The problem is a multi-class classification problem, and we will model it using a softmax activation function on the output layer. This means that the model will predict a vector with three elements with the probability that the sample belongs to each of the three classes. Therefore, we must one hot encode the class values before we split the rows into the train and test datasets. We can do this using the Keras to_categorical() function.
|
1 2 3 4 5 6 7 8 9 |
# generate 2d classification dataset X, y = make_blobs(n_samples=1100, centers=3, n_features=2, cluster_std=2, random_state=2) # one hot encode output variable y = to_categorical(y) # split into train and test n_train = 100 trainX, testX = X[:n_train, :], X[n_train:, :] trainy, testy = y[:n_train], y[n_train:] print(trainX.shape, testX.shape) |
Next, we can define and combine the model.
The model will expect samples with two input variables. The model then has a single hidden layer with 25 nodes and a rectified linear activation function, then an output layer with three nodes to predict the probability of each of the three classes and a softmax activation function.
Because the problem is multi-class, we will use the categorical cross entropy loss function to optimize the model and the efficient Adam flavor of stochastic gradient descent.
|
1 2 3 4 5 |
# define model model = Sequential() model.add(Dense(25, input_dim=2, activation='relu')) model.add(Dense(3, activation='softmax')) model.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy']) |
The model is fit for 500 training epochs and we will evaluate the model each epoch on the test set, using the test set as a validation set.
|
1 2 |
# fit model history = model.fit(trainX, trainy, validation_data=(testX, testy), epochs=500, verbose=0) |
At the end of the run, we will evaluate the performance of the model on the train and test sets.
|
1 2 3 4 |
# evaluate the model _, train_acc = model.evaluate(trainX, trainy, verbose=0) _, test_acc = model.evaluate(testX, testy, verbose=0) print('Train: %.3f, Test: %.3f' % (train_acc, test_acc)) |
Then finally, we will plot learning curves of the model accuracy over each training epoch on both the training and validation datasets.
|
1 2 3 4 5 |
# learning curves of model accuracy pyplot.plot(history.history['accuracy'], label='train') pyplot.plot(history.history['val_accuracy'], label='test') pyplot.legend() pyplot.show() |
Tying all of this together, the complete example is listed below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 |
# develop an mlp for blobs dataset from sklearn.datasets import make_blobs from keras.utils import to_categorical from keras.models import Sequential from keras.layers import Dense from matplotlib import pyplot # generate 2d classification dataset X, y = make_blobs(n_samples=1100, centers=3, n_features=2, cluster_std=2, random_state=2) # one hot encode output variable y = to_categorical(y) # split into train and test n_train = 100 trainX, testX = X[:n_train, :], X[n_train:, :] trainy, testy = y[:n_train], y[n_train:] print(trainX.shape, testX.shape) # define model model = Sequential() model.add(Dense(25, input_dim=2, activation='relu')) model.add(Dense(3, activation='softmax')) model.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy']) # fit model history = model.fit(trainX, trainy, validation_data=(testX, testy), epochs=500, verbose=0) # evaluate the model _, train_acc = model.evaluate(trainX, trainy, verbose=0) _, test_acc = model.evaluate(testX, testy, verbose=0) print('Train: %.3f, Test: %.3f' % (train_acc, test_acc)) # learning curves of model accuracy pyplot.plot(history.history['accuracy'], label='train') pyplot.plot(history.history['val_accuracy'], label='test') pyplot.legend() pyplot.show() |
Running the example first prints the shape of each dataset for confirmation, then the performance of the final model on the train and test datasets.
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
In this case, we can see that the model achieved about 85% accuracy on the training dataset, which we know is optimistic, and about 80% on the test dataset, which we would expect to be more realistic.
|
1 2 |
(100, 2) (1000, 2) Train: 0.850, Test: 0.809 |
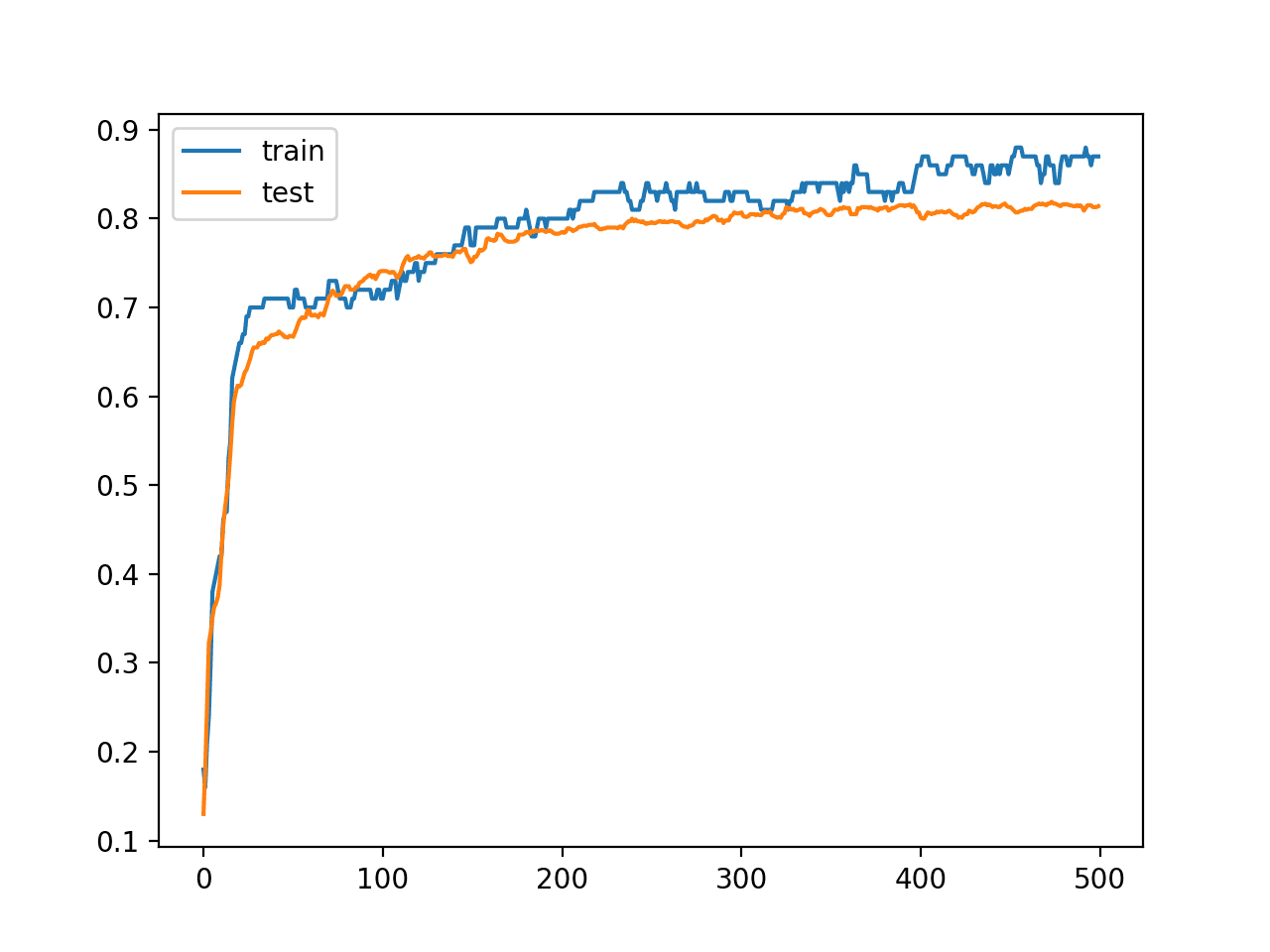
A line plot is also created showing the learning curves for the model accuracy on the train and test sets over each training epoch.
We can see that training accuracy is more optimistic over most of the run as we also noted with the final scores.
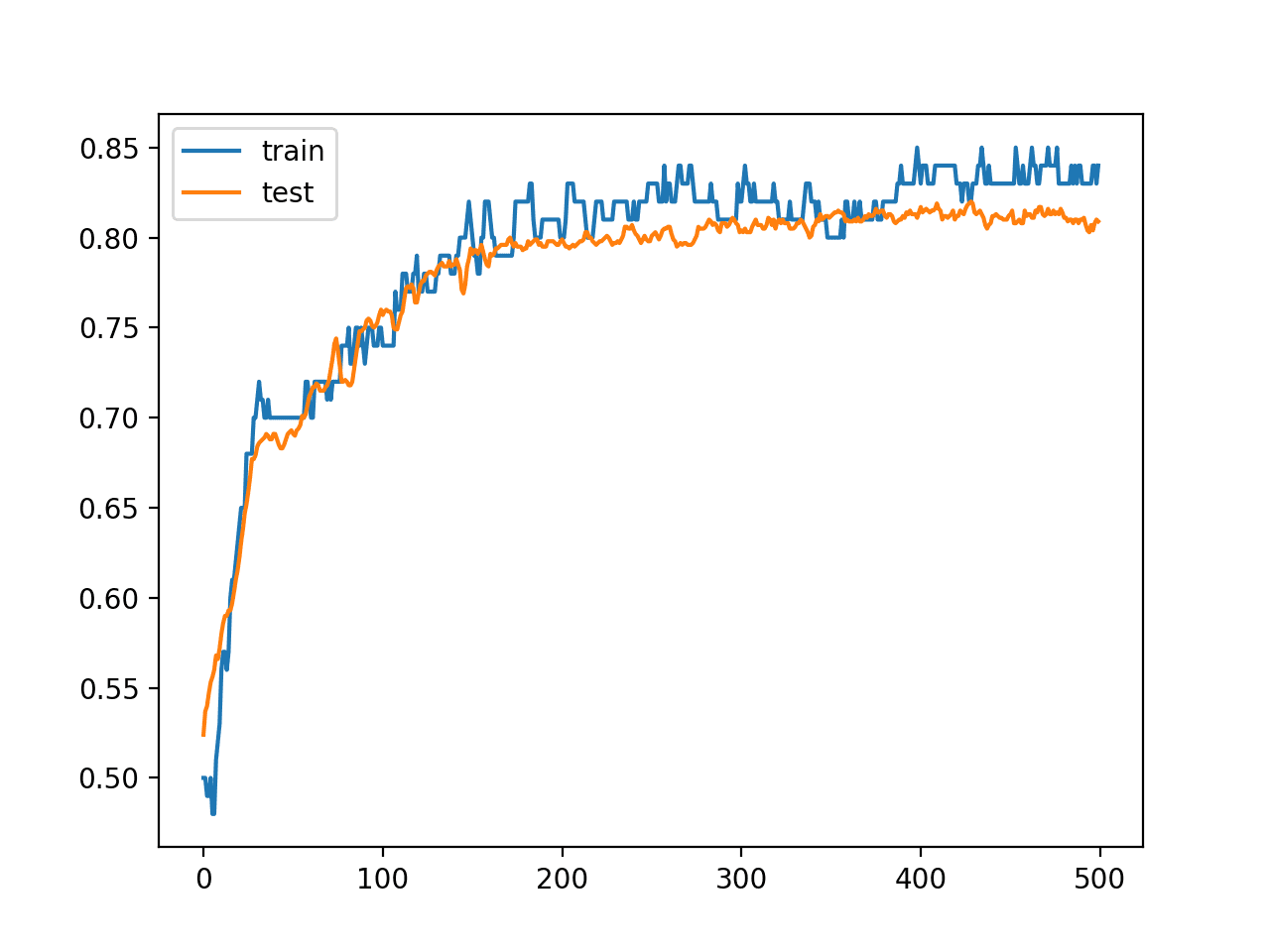
Line Plot Learning Curves of Model Accuracy on Train and Test Dataset Over Each Training Epoch
We can now look at using instances of this model as part of a stacking ensemble.
Train and Save Sub-Models
To keep this example simple, we will use multiple instances of the same model as level-0 or sub-models in the stacking ensemble.
We will also use a holdout validation dataset to train the level-1 or meta-learner in the ensemble.
A more advanced example may use different types of MLP models (deeper, wider, etc.) as sub-models and train the meta-learner using k-fold cross-validation.
In this section, we will train multiple sub-models and save them to file for later use in our stacking ensembles.
The first step is to create a function that will define and fit an MLP model on the training dataset.
|
1 2 3 4 5 6 7 8 9 10 |
# fit model on dataset def fit_model(trainX, trainy): # define model model = Sequential() model.add(Dense(25, input_dim=2, activation='relu')) model.add(Dense(3, activation='softmax')) model.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy']) # fit model model.fit(trainX, trainy, epochs=500, verbose=0) return model |
Next, we can create a sub-directory to store the models.
Note, if the directory already exists, you may have to delete it when re-running this code.
|
1 2 |
# create directory for models makedirs('models') |
Finally, we can create multiple instances of the MLP and save each to the “models/” subdirectory with a unique filename.
In this case, we will create five sub-models, but you can experiment with a different number of models and see how it impacts model performance.
|
1 2 3 4 5 6 7 8 9 |
# fit and save models n_members = 5 for i in range(n_members): # fit model model = fit_model(trainX, trainy) # save model filename = 'models/model_' + str(i + 1) + '.h5' model.save(filename) print('>Saved %s' % filename) |
We can tie all of these elements together; the complete example of training the sub-models and saving them to file is listed below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 |
# example of saving sub-models for later use in a stacking ensemble from sklearn.datasets import make_blobs from keras.utils import to_categorical from keras.models import Sequential from keras.layers import Dense from matplotlib import pyplot from os import makedirs # fit model on dataset def fit_model(trainX, trainy): # define model model = Sequential() model.add(Dense(25, input_dim=2, activation='relu')) model.add(Dense(3, activation='softmax')) model.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy']) # fit model model.fit(trainX, trainy, epochs=500, verbose=0) return model # generate 2d classification dataset X, y = make_blobs(n_samples=1100, centers=3, n_features=2, cluster_std=2, random_state=2) # one hot encode output variable y = to_categorical(y) # split into train and test n_train = 100 trainX, testX = X[:n_train, :], X[n_train:, :] trainy, testy = y[:n_train], y[n_train:] print(trainX.shape, testX.shape) # create directory for models makedirs('models') # fit and save models n_members = 5 for i in range(n_members): # fit model model = fit_model(trainX, trainy) # save model filename = 'models/model_' + str(i + 1) + '.h5' model.save(filename) print('>Saved %s' % filename) |
Running the example creates the “models/” subfolder and saves five trained models with unique filenames.
|
1 2 3 4 5 6 |
(100, 2) (1000, 2) >Saved models/model_1.h5 >Saved models/model_2.h5 >Saved models/model_3.h5 >Saved models/model_4.h5 >Saved models/model_5.h5 |
Next, we can look at training a meta-learner to make best use of the predictions from these submodels.
Separate Stacking Model
We can now train a meta-learner that will best combine the predictions from the sub-models and ideally perform better than any single sub-model.
The first step is to load the saved models.
We can use the load_model() Keras function and create a Python list of loaded models.
|
1 2 3 4 5 6 7 8 9 10 11 12 |
# load models from file def load_all_models(n_models): all_models = list() for i in range(n_models): # define filename for this ensemble filename = 'models/model_' + str(i + 1) + '.h5' # load model from file model = load_model(filename) # add to list of members all_models.append(model) print('>loaded %s' % filename) return all_models |
We can call this function to load our five saved models from the “models/” sub-directory.
|
1 2 3 4 |
# load all models n_members = 5 members = load_all_models(n_members) print('Loaded %d models' % len(members)) |
It would be useful to know how well the single models perform on the test dataset as we would expect a stacking model to perform better.
We can easily evaluate each single model on the training dataset and establish a baseline of performance.
|
1 2 3 4 5 |
# evaluate standalone models on test dataset for model in members: testy_enc = to_categorical(testy) _, acc = model.evaluate(testX, testy_enc, verbose=0) print('Model Accuracy: %.3f' % acc) |
Next, we can train our meta-learner. This requires two steps:
- Prepare a training dataset for the meta-learner.
- Use the prepared training dataset to fit a meta-learner model.
We will prepare a training dataset for the meta-learner by providing examples from the test set to each of the submodels and collecting the predictions. In this case, each model will output three predictions for each example for the probabilities that a given example belongs to each of the three classes. Therefore, the 1,000 examples in the test set will result in five arrays with the shape [1000, 3].
We can combine these arrays into a three-dimensional array with the shape [1000, 5, 3] by using the dstack() NumPy function that will stack each new set of predictions.
As input for a new model, we will require 1,000 examples with some number of features. Given that we have five models and each model makes three predictions per example, then we would have 15 (3 x 5) features for each example provided to the submodels. We can transform the [1000, 5, 3] shaped predictions from the sub-models into a [1000, 15] shaped array to be used to train a meta-learner using the reshape() NumPy function and flattening the final two dimensions. The stacked_dataset() function implements this step.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 |
# create stacked model input dataset as outputs from the ensemble def stacked_dataset(members, inputX): stackX = None for model in members: # make prediction yhat = model.predict(inputX, verbose=0) # stack predictions into [rows, members, probabilities] if stackX is None: stackX = yhat else: stackX = dstack((stackX, yhat)) # flatten predictions to [rows, members x probabilities] stackX = stackX.reshape((stackX.shape[0], stackX.shape[1]*stackX.shape[2])) return stackX |
Once prepared, we can use this input dataset along with the output, or y part, of the test set to train a new meta-learner.
In this case, we will train a simple logistic regression algorithm from the scikit-learn library.
Logistic regression only supports binary classification, although the implementation of logistic regression in scikit-learn in the LogisticRegression class supports multi-class classification (more than two classes) using a one-vs-rest scheme. The function fit_stacked_model() below will prepare the training dataset for the meta-learner by calling the stacked_dataset() function, then fit a logistic regression model that is then returned.
|
1 2 3 4 5 6 7 8 |
# fit a model based on the outputs from the ensemble members def fit_stacked_model(members, inputX, inputy): # create dataset using ensemble stackedX = stacked_dataset(members, inputX) # fit standalone model model = LogisticRegression() model.fit(stackedX, inputy) return model |
We can call this function and pass in the list of loaded models and the training dataset.
|
1 2 |
# fit stacked model using the ensemble model = fit_stacked_model(members, testX, testy) |
Once fit, we can use the stacked model, including the members and the meta-learner, to make predictions on new data.
This can be achieved by first using the sub-models to make an input dataset for the meta-learner, e.g. by calling the stacked_dataset() function, then making a prediction with the meta-learner. The stacked_prediction() function below implements this.
|
1 2 3 4 5 6 7 |
# make a prediction with the stacked model def stacked_prediction(members, model, inputX): # create dataset using ensemble stackedX = stacked_dataset(members, inputX) # make a prediction yhat = model.predict(stackedX) return yhat |
We can use this function to make a prediction on new data; in this case, we can demonstrate it by making predictions on the test set.
|
1 2 3 4 |
# evaluate model on test set yhat = stacked_prediction(members, model, testX) acc = accuracy_score(testy, yhat) print('Stacked Test Accuracy: %.3f' % acc) |
Tying all of these elements together, the complete example of fitting a linear meta-learner for the stacking ensemble of MLP sub-models is listed below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 |
# stacked generalization with linear meta model on blobs dataset from sklearn.datasets import make_blobs from sklearn.metrics import accuracy_score from sklearn.linear_model import LogisticRegression from keras.models import load_model from keras.utils import to_categorical from numpy import dstack # load models from file def load_all_models(n_models): all_models = list() for i in range(n_models): # define filename for this ensemble filename = 'models/model_' + str(i + 1) + '.h5' # load model from file model = load_model(filename) # add to list of members all_models.append(model) print('>loaded %s' % filename) return all_models # create stacked model input dataset as outputs from the ensemble def stacked_dataset(members, inputX): stackX = None for model in members: # make prediction yhat = model.predict(inputX, verbose=0) # stack predictions into [rows, members, probabilities] if stackX is None: stackX = yhat else: stackX = dstack((stackX, yhat)) # flatten predictions to [rows, members x probabilities] stackX = stackX.reshape((stackX.shape[0], stackX.shape[1]*stackX.shape[2])) return stackX # fit a model based on the outputs from the ensemble members def fit_stacked_model(members, inputX, inputy): # create dataset using ensemble stackedX = stacked_dataset(members, inputX) # fit standalone model model = LogisticRegression() model.fit(stackedX, inputy) return model # make a prediction with the stacked model def stacked_prediction(members, model, inputX): # create dataset using ensemble stackedX = stacked_dataset(members, inputX) # make a prediction yhat = model.predict(stackedX) return yhat # generate 2d classification dataset X, y = make_blobs(n_samples=1100, centers=3, n_features=2, cluster_std=2, random_state=2) # split into train and test n_train = 100 trainX, testX = X[:n_train, :], X[n_train:, :] trainy, testy = y[:n_train], y[n_train:] print(trainX.shape, testX.shape) # load all models n_members = 5 members = load_all_models(n_members) print('Loaded %d models' % len(members)) # evaluate standalone models on test dataset for model in members: testy_enc = to_categorical(testy) _, acc = model.evaluate(testX, testy_enc, verbose=0) print('Model Accuracy: %.3f' % acc) # fit stacked model using the ensemble model = fit_stacked_model(members, testX, testy) # evaluate model on test set yhat = stacked_prediction(members, model, testX) acc = accuracy_score(testy, yhat) print('Stacked Test Accuracy: %.3f' % acc) |
Running the example first loads the sub-models into a list and evaluates the performance of each.
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
We can see that the best performing model is the final model with an accuracy of about 81.3%.
|
1 2 3 4 5 6 7 8 9 10 11 12 |
(100, 2) (1000, 2) >loaded models/model_1.h5 >loaded models/model_2.h5 >loaded models/model_3.h5 >loaded models/model_4.h5 >loaded models/model_5.h5 Loaded 5 models Model Accuracy: 0.805 Model Accuracy: 0.806 Model Accuracy: 0.804 Model Accuracy: 0.809 Model Accuracy: 0.813 |
Next, a logistic regression meta-learner is trained on the predicted probabilities from each sub-model on the test set, then the entire stacking model is evaluated on the test set.
We can see that in this case, the meta-learner out-performed each of the sub-models on the test set, achieving an accuracy of about 82.4%.
|
1 |
Stacked Test Accuracy: 0.824 |
Integrated Stacking Model
When using neural networks as sub-models, it may be desirable to use a neural network as a meta-learner.
Specifically, the sub-networks can be embedded in a larger multi-headed neural network that then learns how to best combine the predictions from each input sub-model. It allows the stacking ensemble to be treated as a single large model.
The benefit of this approach is that the outputs of the submodels are provided directly to the meta-learner. Further, it is also possible to update the weights of the submodels in conjunction with the meta-learner model, if this is desirable.
This can be achieved using the Keras functional interface for developing models.
After the models are loaded as a list, a larger stacking ensemble model can be defined where each of the loaded models is used as a separate input-head to the model. This requires that all of the layers in each of the loaded models be marked as not trainable so the weights cannot be updated when the new larger model is being trained. Keras also requires that each layer has a unique name, therefore the names of each layer in each of the loaded models will have to be updated to indicate to which ensemble member they belong.
|
1 2 3 4 5 6 7 8 |
# update all layers in all models to not be trainable for i in range(len(members)): model = members[i] for layer in model.layers: # make not trainable layer.trainable = False # rename to avoid 'unique layer name' issue layer._name = 'ensemble_' + str(i+1) + '_' + layer.name |
Once the sub-models have been prepared, we can define the stacking ensemble model.
The input layer for each of the sub-models will be used as a separate input head to this new model. This means that k copies of any input data will have to be provided to the model, where k is the number of input models, in this case, 5.
The outputs of each of the models can then be merged. In this case, we will use a simple concatenation merge, where a single 15-element vector will be created from the three class-probabilities predicted by each of the 5 models.
We will then define a hidden layer to interpret this “input” to the meta-learner and an output layer that will make its own probabilistic prediction. The define_stacked_model() function below implements this and will return a stacked generalization neural network model given a list of trained sub-models.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 |
# define stacked model from multiple member input models def define_stacked_model(members): # update all layers in all models to not be trainable for i in range(len(members)): model = members[i] for layer in model.layers: # make not trainable layer.trainable = False # rename to avoid 'unique layer name' issue layer._name = 'ensemble_' + str(i+1) + '_' + layer.name # define multi-headed input ensemble_visible = [model.input for model in members] # concatenate merge output from each model ensemble_outputs = [model.output for model in members] merge = concatenate(ensemble_outputs) hidden = Dense(10, activation='relu')(merge) output = Dense(3, activation='softmax')(hidden) model = Model(inputs=ensemble_visible, outputs=output) # plot graph of ensemble plot_model(model, show_shapes=True, to_file='model_graph.png') # compile model.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy']) return model |
A plot of the network graph is created when this function is called to give an idea of how the ensemble model fits together.
|
1 2 |
# define ensemble model stacked_model = define_stacked_model(members) |
Creating the plot requires that pygraphviz is installed.
If this is a challenge on your workstation, you can comment out the call to the plot_model() function.
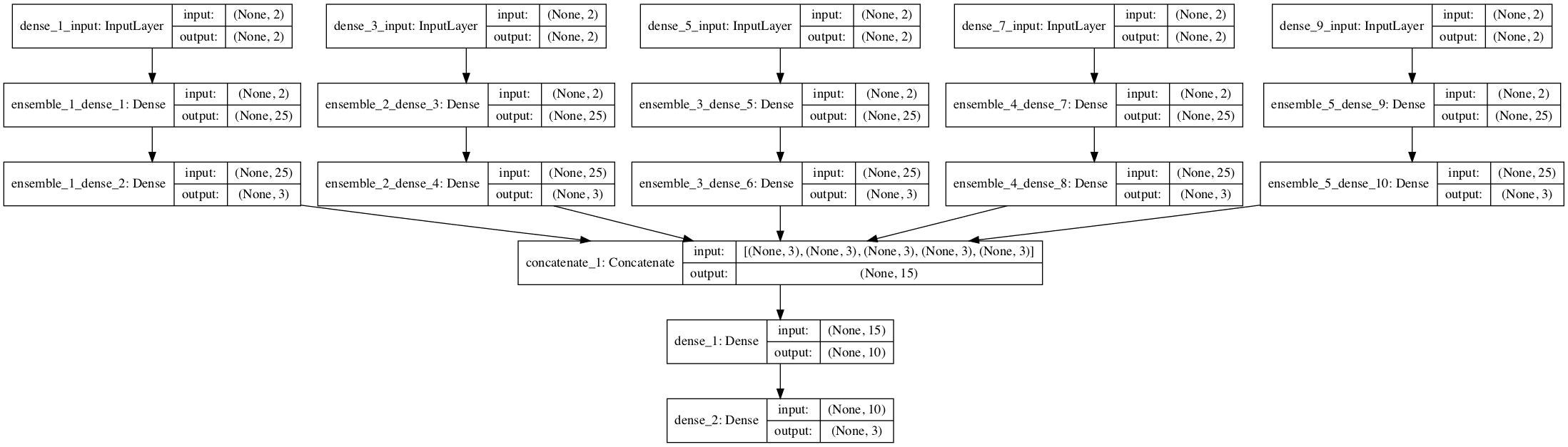
Visualization of Stacked Generalization Ensemble of Neural Network Models
Once the model is defined, it can be fit. We can fit it directly on the holdout test dataset.
Because the sub-models are not trainable, their weights will not be updated during training and only the weights of the new hidden and output layer will be updated. The fit_stacked_model() function below will fit the stacking neural network model on for 300 epochs.
|
1 2 3 4 5 6 7 8 |
# fit a stacked model def fit_stacked_model(model, inputX, inputy): # prepare input data X = [inputX for _ in range(len(model.input))] # encode output data inputy_enc = to_categorical(inputy) # fit model model.fit(X, inputy_enc, epochs=300, verbose=0) |
We can call this function providing the defined stacking model and the test dataset.
|
1 2 |
# fit stacked model on test dataset fit_stacked_model(stacked_model, testX, testy) |
Once fit, we can use the new stacked model to make a prediction on new data.
This is as simple as calling the predict() function on the model. One minor change is that we require k copies of the input data in a list to be provided to the model for each of the k sub-models. The predict_stacked_model() function below simplifies this process of making a prediction with the stacking model.
|
1 2 3 4 5 6 |
# make a prediction with a stacked model def predict_stacked_model(model, inputX): # prepare input data X = [inputX for _ in range(len(model.input))] # make prediction return model.predict(X, verbose=0) |
We can call this function to make a prediction for the test dataset and report the accuracy.
We would expect the performance of the neural network learner to be better than any individual submodel and perhaps competitive with the linear meta-learner used in the previous section.
|
1 2 3 4 5 |
# make predictions and evaluate yhat = predict_stacked_model(stacked_model, testX) yhat = argmax(yhat, axis=1) acc = accuracy_score(testy, yhat) print('Stacked Test Accuracy: %.3f' % acc) |
Tying all of these elements together, the complete example is listed below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 |
# stacked generalization with neural net meta model on blobs dataset from sklearn.datasets import make_blobs from sklearn.metrics import accuracy_score from keras.models import load_model from keras.utils import to_categorical from keras.utils import plot_model from keras.models import Model from keras.layers import Input from keras.layers import Dense from keras.layers.merge import concatenate from numpy import argmax # load models from file def load_all_models(n_models): all_models = list() for i in range(n_models): # define filename for this ensemble filename = 'models/model_' + str(i + 1) + '.h5' # load model from file model = load_model(filename) # add to list of members all_models.append(model) print('>loaded %s' % filename) return all_models # define stacked model from multiple member input models def define_stacked_model(members): # update all layers in all models to not be trainable for i in range(len(members)): model = members[i] for layer in model.layers: # make not trainable layer.trainable = False # rename to avoid 'unique layer name' issue layer._name = 'ensemble_' + str(i+1) + '_' + layer.name # define multi-headed input ensemble_visible = [model.input for model in members] # concatenate merge output from each model ensemble_outputs = [model.output for model in members] merge = concatenate(ensemble_outputs) hidden = Dense(10, activation='relu')(merge) output = Dense(3, activation='softmax')(hidden) model = Model(inputs=ensemble_visible, outputs=output) # plot graph of ensemble plot_model(model, show_shapes=True, to_file='model_graph.png') # compile model.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy']) return model # fit a stacked model def fit_stacked_model(model, inputX, inputy): # prepare input data X = [inputX for _ in range(len(model.input))] # encode output data inputy_enc = to_categorical(inputy) # fit model model.fit(X, inputy_enc, epochs=300, verbose=0) # make a prediction with a stacked model def predict_stacked_model(model, inputX): # prepare input data X = [inputX for _ in range(len(model.input))] # make prediction return model.predict(X, verbose=0) # generate 2d classification dataset X, y = make_blobs(n_samples=1100, centers=3, n_features=2, cluster_std=2, random_state=2) # split into train and test n_train = 100 trainX, testX = X[:n_train, :], X[n_train:, :] trainy, testy = y[:n_train], y[n_train:] print(trainX.shape, testX.shape) # load all models n_members = 5 members = load_all_models(n_members) print('Loaded %d models' % len(members)) # define ensemble model stacked_model = define_stacked_model(members) # fit stacked model on test dataset fit_stacked_model(stacked_model, testX, testy) # make predictions and evaluate yhat = predict_stacked_model(stacked_model, testX) yhat = argmax(yhat, axis=1) acc = accuracy_score(testy, yhat) print('Stacked Test Accuracy: %.3f' % acc) |
Running the example first loads the five sub-models.
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
A larger stacking ensemble neural network is defined and fit on the test dataset, then the new model is used to make a prediction on the test dataset. We can see that, in this case, the model achieved an accuracy of about 83.3%, out-performing the linear model from the previous section.
|
1 2 3 4 5 6 7 8 |
(100, 2) (1000, 2) >loaded models/model_1.h5 >loaded models/model_2.h5 >loaded models/model_3.h5 >loaded models/model_4.h5 >loaded models/model_5.h5 Loaded 5 models Stacked Test Accuracy: 0.833 |
Extensions
This section lists some ideas for extending the tutorial that you may wish to explore.
- Alternate Meta-Learner. Update the example to use an alternate meta-learner classifier model to the logistic regression model.
- Single Level 0 Model. Update the example to use a single level-0 model and compare the results.
- Vary Level 0 Models. Develop a study that demonstrates the relationship between test classification accuracy and the number of sub-models used in the stacked ensemble.
- Cross-Validation Stacking Ensemble. Update the example to use k-fold cross-validation to prepare the training dataset for the meta-learner model.
- Use Raw Input in Meta-Learner. Update the example so that the meta-learner algorithms take the raw input data for the sample as well as the output from the sub-models and compare performance.
If you explore any of these extensions, I’d love to know.
Further Reading
This section provides more resources on the topic if you are looking to go deeper.
Books
- Section 8.8 Model Averaging and Stacking, The Elements of Statistical Learning: Data Mining, Inference, and Prediction, Second Edition, 2016.
- Section 7.5 Combining multiple models, Data Mining: Practical Machine Learning Tools and Techniques, Second Edition, 2005.
- Section 9.8.2 Stacked Generalization, Neural Networks for Pattern Recognition, 1995.
Papers
- Stacked generalization, 1992.
- Issues in Stacked Generalization, 1999.
API
- Getting started with the Keras Sequential model
- Keras Core Layers API
- numpy.argmax API
- sklearn.datasets.make_blobs API
- numpy.dstack API
- sklearn.linear_model.LogisticRegression API
Articles
Posts
Summary
In this tutorial, you discovered how to develop a stacked generalization ensemble for deep learning neural networks.
Specifically, you learned:
- Stacked generalization is an ensemble method where a new model learns how to best combine the predictions from multiple existing models.
- How to develop a stacking model using neural networks as a submodel and a scikit-learn classifier as the meta-learner.
- How to develop a stacking model where neural network sub-models are embedded in a larger stacking ensemble model for training and prediction.
Do you have any questions?
Ask your questions in the comments below and I will do my best to answer.





Hi Jason,
Awesome post. I tried to perform a stacking ensemble for a binary classification task. However, I have an issue with the stacked_dataset definition. It gives a value error saying “bad_shape (200, 2)”. I have 200 test samples with 2 classes. Kindly suggest the modification needed for the stacked_dataset definition. Many thanks.
You may have to adapt the example to your specific models and dataset.
Hi Sivarama,
I had the same problem here. The thing was that I set the one-hot encoded list (testy_enc) to the fit_stacked_model function, which calls stacked_dataset later, but it should be the binary list (testy) instead.
Raniery
Thank you
No problem.
Tableau
What about it?
Since you uses multiple same height models, is it possible to use different sub models like different layers or different type, VGG, Inception etc. as the sub models?
Another question is, if we shuffle the data sets between the sub models, how possible the stacking ensemble model over fitting?
Yes, you can use models of varying depth.
Fitting each model on separate data would no longer be a stacking model, but instead a bagging type model.
Hi Jason,
Thank you for your tutorial. It was very helpful!
A question for you: Is it possible to stack models using the same algorithms, but with different data types (i.e. the level 0 models are two logistic regression models – one using numerical data and one with text, and the outputs of both are stacked into a single level 1 logistic regression model?)
Thank you,
Yes!
Thank you Jason. Although I need to digest all you have written as I am a newbie this this field, I appreciate your effort in sharing your knowledge.
Thanks. Stick with it!
Thank you Jason.
I also appreciate your effort in sharing your knowledge.
Thanks.
Thanks for this article, Jason. I tried the extension “Use Raw Input in Meta-Learner.” My understanding is that you take the output of the 0-level models, and then join that to the data that were used to make the output (predictions) of the 0-level models. So the input to the meta-model would be (n,X+Y), where n is the number of observations in the validation set, X is the number of features in the raw data, and Y is the number of 0-level models (for a binary classification problem). My intuition would be that this is over fitting, and it looks like it may be, based on the meta-model performance. Wouldn’t the meta-model then be getting “information” from that raw data twice? Once indirectly through the 0-level models, and then again through the raw data? Or is this legit because the “raw data” being provided to the meta-model was only used to validate, not train, the 0-level models?
Thanks again for your article (and all your other ones!) I find them very useful.
The input would be the input to the level 0 models (X) and the output from each level 0 model (yhats).
The raw input (X) provides additional context for the level 0 outputs (yhats)
It may or may not overfit. Often it results in better performance.
Hi Jason,
Thank you for the interesting post. I have a quick question on the accuracy scores between base-learners and meta-learn.
For base-learners, you used (trainX, tainY) to train them, and evaluate acc on (testX, testY).
For meta-learners, you used (testX, testY) to train them, and evaluate acc on (testX, testY) again. Wouldn’t the acc for meta-learners be inflated? Would it be better to have a ‘true hold-out set’ that none of the base- and meta-learner have seen?
Thanks
Yes, that would be a nice improvement.
thank you sir , can you suggest some machine learning algorithms using MATLAB, particularly regarding Reproducing Hilbert kernel space and implementation of kernel trick .
Sorry, I don’t have any matlab examples.
A great post indeed. What I don’t fully understand is, why the test data is used to fit the stacked model, and then the stacked model is evaluated against the test data? Isn’t it overfitting the ensemble model?
Another error I happened to have, is when I loading models some of them happened to have the same layer names. The renaming function however doesn’t work with the input layers, so the code of ‘Model(inputs=ensemble_inputs…)’ throws an error saying there are duplicates in input layer names.
Yes, that is the clever part. Each round uses a different holdout set to find the coefficients for combining the models – together all data was unseen in the estimation of the coefficients. It should not overfit, ideally.
Perhaps double check that your version of Keras is up to date.
Just to clarify what do you mean by ‘each round’, as in the last example where the NN is used as a meta-learner, there is no cross-validation deployed. I understand the sub-models are stacked together and the training is done at one go.
And I am using the latest Keras 2.2.4 version.
In that case, we load the pre-trained models and fit the data using a “new” dataset not used to train the submodels, e.g. in this case the test dataset.
Ideally, we would not use the test dataset and instead use a subset of the train dataset not seen by the submodels.
Agree. So in this example above, the stacked model is trained on test dataset and then evaluated against test dataset isn’t ideal and potentially overfits the test dataset?
Yes, correct.
Thanks for the great tutorial. As Oliver asked you, I didn’t understand why you used same dataset (testX) for fitting and predicting meta-model. Let’s say I have training dataset and test dataset and I just have labels for training dataset not test dataset. So I am not able to use test data set for fitting and just can use it for predicting. So should I divide training dataset and use the unseen part for fitting meta-model?
Thanks,
For brevity, ideally you would fit the meta model on a validation dataset.
Train base models on one set (train), then meta model on another set (val), then evaluate in another set (test).
Then could you please clarify it in the post as it’s misleading to readers. The cross-validation shouldn’t be optional in this case but a must in order to generate inputs to the meta-learner. If there are N rows in the training set and L sub-models, the cross-validation should first produce a N * L matrix, together with the y_train to train the meta-learner.
That means a cross-validation needs to be implemented in the ‘fit_stacked_model’ function, and input trainX and trainY instead.
Thanks for the suggestion.
My pleasure. Thanks for the great post. Keep it up.
Hi Jason,
thanks for the great post! I would like to use it to stack my own neural networks. They are also Sequential models – but I get the following issue: Two of the input tensors have the same name. And as tensors can have no name change, the code you provided to change the layer names doesn’t alter them. Hence I get the error:
ValueError: The name “dense_7_input” is used 2 times in the model. All layer names should be unique.
I have tried quite a few things – but no success. However, I don’t really care about the actual models, I could retrain them and alter the name of the input while setting them up. But I have found no way to give the input tensor created within the input layer of a Sequential model a custom name.
What I can’t do though is to create all five (yes, I also have five models to concatenate, by chance) in one single loop as training one of them takes up to several hours. Have you got any suggestions how I could find a workaround? I would be very greatful.
best regards and thanks a lot again and in advance,
Wolfgang
You can specify the name of layers when the layer is created via the name attribute.
You can also alter the name attribute of a layer after it is loaded, then save it again.
Hi Jason,
thanks a lot for your help. I couldn’t change the name of the attribute – but I sorted it differently. If I save the weights of each model, set up a new model from scratch (with explicitly naming the first layer, i.e. not renaming layers in a loop) and then add the weights to the reloaded model. It works, I am now testing it. Thanks again and best regards, Wolfgang
Nice workaround, thanks for sharing.
Hi Jason
thanks for your comment – and sorry to bother you again. I have another question and I wonder whether you have any “high level” tip as to where I could look up what to do.
For my project (in computer vision) I have five feed forward nets with features extracted of images – and one (later maybe two or three) CNNs.
I could concatenate the feed forward nets and the overall accuracy, basically using the code of your blog, increases by about 1.5 percent in comparison to my previous method of combining the output of each feed forward net. Previously I averaged the softmax probabilities (see Geoffrey Hintons course on Coursera, lecture 10 for more details), finding the best weight for each net with a brute force approach on the saved softmax output of the validation set, so that the model is not optimized on the test set.
However, if I average the softmax output probability predictions of the stacked model with my CNN softmax output probabilities, the overall accuracy drops by about two percent (even though the stacked feed forward accuracy is better than before) with respect to my previous method. Apart from the fact that I wonder why this is, I thought maybe I could add the CNN to the stacked model and see whether that improves the overall accuracy.
The problem is: For the CNN I have to (for memory reasons) use a data generator – and I don’t know how I could set up a stacked model with that.
Do you have any clues whether stacking a CNN and some feed forward nets, the first using a data generator and the others not, is possible at all? And if so – do you have a reference page, a blog post or something like it where I could look it up?
My second question is related to that: Could I use the saved output probabilities of each net to set up a stacked neural network (as this would get around the data generator problem).
Thanks again for your kind help,
best regards,
Wolfgang
Hmmm, if each model takes the same input, then each model can use the same data generator.
Otherwise, you may have to write custom code to generate samples and feed them through your models sample by sample – I guess. Some prototyping might be required.
Yes, you can use saved out of sample predictions to fit a stacked model or a weighting for predictions from multiple models, e.g.:
https://machinelearningmastery.com/weighted-average-ensemble-for-deep-learning-neural-networks/
Thanks for this post!
I still got the ‘unique layer name’ error message even though I set the new layer names on each level 0 model as you did above. I didn’t almost changed your code.
The error message was like this
—————————————-
ValueError: The name “dense_1_input” is used 5 times in the model. All layer names should be unique.
—————————————-
But when I tried to find layers that have that name none of the layers has that name
—————————————-
model.get_layer(“dense_1_input”)
>>> ValueError: No such layer: dense_1_input
—————————————-
Now I got it! We should add one more line.
——————–
model.input_names = ‘ensemble_’ + str(i+1) + ‘_’ + model.input_names
——————–
OMG that didn’t work……………………
Hang in there, I’m sure it is a small bug that once fixed will fix your whole model.
Perhaps summarize (e.g. model.summary()) the model to confirm the layer names are indeed different prior to stacking?
Already check that, the names were all changed but I got still the same error.
I want to ask you another question, If I do 10 fold cross validation then I’d get 10 (level 0) models. To make predictions on test set (unseen data) at the last step, which model among the 10 (level 0) models should I feed the testX to?
If I used 5 (level 0) models based on a holdout dataset, then in the 10-fold CV case the total # of models would be 5*10, right? and after feeding the test set to the all 10 pairs I’d get 10 times more many yhat.. (but actual y values can’t be prepared 10 times more many)
I’m not sure I follow, why would you feed testX to model trained on 9/10s of your data?
Because the level 1 model was trained by the data that is fed through the level 0 models? So if I want to make predictions on the test set, I have to feed the test set to the level 0 models and then to the level 1 model sequentially. Is this correct?
Yes, but there is no k-fold split involved.
Hi I’m getting this error also because the error comes from the model’s type and not the name. The format in the model.summary() is:
layerName (layerType)
ensemble_1_cu_dnnlstm_1 (CuDNNLSTM)
so we see that only the name changed but the type is still CuDNNLSTM which will raise the error.
Were you able to find a fix for this error? 🙁
I don’t know why you are getting this error, sorry.
Perhaps try using LSTMs, confirm it works, then change to Cuda LSTMs.
Hi Jason! Thank you for sharing your approach. I’m trying to find the best way to solve my problem and will really appreciate your suggestions. I have 3 different NNs doing the same predictions but using different data sets: text, images, and metadata. Would your way of stacking them together with another NN work? Thanks!
Perhaps try it and see how results compare to a simple average of the three models?
What should I pass to the final model as testX in
fit_stacked_model(stacked_model, testX, testy)in my case?Hi, Jason. It’s very informative contents.
I have a question about the shape of stackX. Isn’t it [1000, 3, 5] with stackX shape although you mentioned [1000, 5, 3] in stacked_dataset function? Because I understand as [1000,3] *5 (yhat * 5 models).
Yes, this is covered in the section “Separate Stacking Model”.
The shape is [1000, 5, 3] for the 5 models that make a prediction for 3 classes.
Thanks for the tuts!
logistic regression give a value error bad input shape. According to doc, fit() expect y : array-like, shape (n_samples,) but testy here is of shape (1000, 3). What am I missing?
Sorry to hear that, I have some suggestions here:
https://machinelearningmastery.com/faq/single-faq/why-does-the-code-in-the-tutorial-not-work-for-me
Thanks, jason!
This is what I did : I used np.argmax(testy, axis=1) to address the issue. Also in the fit_stacked_model(), the additional categorical encoding seems not be necessary since testy was encoded already before the data splitting step.
Nice work!
Hi Jason,
Sorry to bother you. But when running the define_stacked_model, I get the following error
The name “conv2d_1_input” is used 10 times in the model. All layer names should be unique.
at this step
model = Model(inputs=ensemble_visible, outputs=output)
I’m stacking 10 models, I’ve checked every one of them and the layers names are different because you already made sure to rename them in your code. Any advice on how to fix this?
Thanks.
Yes, note the section where we change the names of the layers in the model.
I believe you may have skipped a section of code.
Hi Jason!
Great informative post on stacking ensembles! I am currently using pretrained models (Inception, VGG, Resnet) and using transfer learning to train the models on specific medical images.
It seems like when I use the models individually, my loss is fairly low(0.0833).
When I try using a stack ensemble however, my loss gets as high as 13!
Do you have an idea what might be the cause of this issue?
I am using an adam optimzer, with a hidden layer of 4096 nodes with an relu activation function. The output layer uses a softmax activation function.
Perhaps there is a bug in the way the models are being combined?
Perhaps look at accuracy rather than loss?
Perhaps start with a simple average or weighted average and go from there?
Hm thats what Im thinking, but for some iterations(using leave one group cross validator) i do get a good low loss. Accuracy is fairly bad throughout all iterations however.
Posting in case this helps anyone!
My high loss rate came from having too many hidden layers (4096) and not enough data. Droping the number from 4096->100 vastly improved my loss rate!
Wow, that is a ton of hidden layers.
Do you mean nodes in a hidden layer?
Hello, very thankful to be able to browse your question. I am also trying to use the method you mentioned above to solve my medical images classification task which has also limited data. Would you mind sharing me your code or how did you do that? My email address is chenxiao97619@gmail.com Hope for your reply. Many thanks.
Hello sir
i loved your tutorials. i am trying and coming your k-fold validation tutorial with this stacking tutorial. for 5-fold validation and 5 base models, when i am trying generating stacked ensemble (with total 25 (5*5) i am getting error for renaming layers.
‘ValueError: The name “embedding_input” is used 3 times in the model. All layer names should be unique.’
i am exactly using the same command as written by you.
for layer in model.layers:
# make not trainable
layer.trainable = False
# rename to avoid ‘unique layer name’ issue
layer._name = ‘ensemble_’ + str(i+1) + ‘_’ + layer.name
What should i do to remove error?
Please help
I applied that concept on a simple CNN for dog breeds predictions (Without applying data augmentation and/or transfer learning) and it got from an average of 20% of accuracy to an insane final model with 51.196 % accuracy, that’s was quite unexpected. I’m thinking that’s too good be true LoL, so I’ll run some validations and then test with the transfer learning model that has around 85%. I’ll share my results once I complete the project.
Excellent article!
I would expect an accuracy close to 100% (high 90s) for a problem like that when using transfer learning and data augmentation.
You got the results?
Thanks,Jason. It’s very useful for me. And I have a question that can I use Pytorch to implement this?
I don’t see why not.
Hi Jason, it’s a very helpful post, I appreciate you. Just a question,
In model = fit_stacked_model(members, testX, testy), you are fitting an LR model to the predictions of sub-models. Then in yhat = stacked_prediction(members, model, testX) you are predicting on the same stacked test data, the fit_stacked_model (meta-learner) shouldn’t be fitted over train stacked predictions?
Yes, I used the same data for brevity.
Ideally, you would fit the base models on train. Then fit the stacked on validation. Then prediction on test.
https://machinelearningmastery.com/faq/single-faq/why-do-you-use-the-test-dataset-as-the-validation-dataset
Thank you for such a great tutorial. I have developed a CNN-LSTM regression model.
The input shape is (None, 58, 2) and output is (None, 1).
1. If i run my model 5 times and save the best model each time. It means i have 5 models ready for ensemble.
2. If i use Integrating Stacking Model concatenate the output shape of those models, the shape will be (None, 5). I can use a fully connected dense layer and output layer to get the final output of the ensemble model.
My question is following:
Can i use the same training data to train the ensemble model? or i need to use different data for training the ensemble model.
Thank you so much.
Irtaza
Nice work.
Good question. Ideally, the ensemble is fit either on a separate holdout dataset.
Hi. Thank you for the tutorial.
I am trying to implement the part where a neural network is the meta learner.
I changed the layer names but when I run
model = Model(inputs=ensemble_visible, outputs=output)
I get an error ValueError: The name “sequential_8_input” is used 2 times in the model. All layer names should be unique.
“sequential_8_input” corresponds to the name of the input tensor and I can’t find a way to change it. Please help.
You may need to change the names of the layers, either when they are defined or after you create the model.
Each layer has a .name property you can set or a name= argument you can set.
Hello,
I am facing the same issue for my stacked model. I have changed every name of the layers but the model.input & model,output have the same name.
@Hajer, did you manage to correct your issue?
Thanks for the tutorial.
Sorry to hear that, perhaps some of these suggestions will help:
https://machinelearningmastery.com/faq/single-faq/why-does-the-code-in-the-tutorial-not-work-for-me
Hi Jason,
With my small neural network, I am performing 5fold CV(30 iterations) 30 times, saving 30 models and then stacking them in the ensemble using separate stacking model(logistic regression).
I run this once and my stacked test accuracy is 87. I run this again and the stacked test accuracy goes down to 84.
Any leads to why the system does not produce reproducible results even after so many iterations and cross-validation models?
Thanks for your recent help as well! I am able to cater for data variance and model variance of NN to some extent.
Nice work.
I have many tutorials on reducing model variance here that may provide a useful starting point:
https://machinelearningmastery.com/start-here/#better
Thanks, Jason. I figured it out. It was some other issue(wasn’t scaling my data on test using just transform). Stable predictions for 4 runs I think, 86.5 ±0.98 (±1.13%). Thank you again for these awesome tutorials 🙂
Really nice work, well done!
Jason, thanks for the tutorial.
I tried to reproduce your example in a little bit different way and faced a problem: Let’s suppose you need to train multiple models, in this case to avoid memory leak, you need to insert a keras.backend.K.clear_session(), the code would be this:
# fit a stacked model
def fit_stacked_model (model, inputX, inputy):
# prepare input data
X = [inputX for _ in range (len (model.input))]
# encode output data
inputy_enc = to_categorical (inputy)
# fit model
model.fit (X, inputy_enc, epochs = 300, verbose = 0)
model.save (‘[NameToSave] .h5’)
K.clear_session ()
When I run
fit_stacked_model (stacked_model, testX, testy)
Everything works perfectly, however if I run again:
fit_stacked_model (stacked_model, testX, testy)
Raises the following error:
TypeError: Cannot interpret feed_dict key as Tensor: Tensor Tensor (“dense_1_input_1: 0”, shape = (?, 2), dtype = float32) is not an element of this graph.
Could you help me find out what am I missing?
Thanks again
Perhaps don’t clear the session?
Hi Jason, thank you for reply me; but if I do not use the clear_session() I will have problems with memmory after run several models
Why do you have problems with memory?
Is it possible that there is something going on with your TF installation?
Hi Jason,
I don’t know why that happens but changing my tensorflow’s version solved the problem, thank you again!
Happy to hear that.
Hi Jason,
Excellent post. I’m missing something quite basic here. You pass the exact same data to the submodel calibration function (i.e., we are not bagging), so what makes the level0 sub-models different from each other exactly? Is there an element of randomness in how the MLPs are calibrated?
Thanks!
Thanks.
I fit the next level model on the same data for brevity, you and should use a separate validation dataset.
I’m comparing the 5 different submodels. Why are they different? Same inputs… Is there a random seed in the adam optimizer?
Because of the stochastic learning algorithm, more here:
https://machinelearningmastery.com/faq/single-faq/why-do-i-get-different-results-each-time-i-run-the-code
Got it – thank you!
Happy to hear that.
Hi, thanks for the post
I have one query i split my data into training and testing
and than trained 3 CNN model using training data. Further the meta learner is trained using the same training data and validated using holdout test data and I am getting good testing results for meta learner as compared to base learners.
is it right way to train the meta learner with same training set and validate with same test set used for level 0 model.
You’re welcome.
Good question.
It is probably best to fit the meta learner to correct the hold out predictions of the base learners. I see two easy ways:
1. fit the base learners on one dataset. Then get another dataset, run the data through the base learners, then use those outputs to fit the meta learner.
2. use one dataset, split into k-folds, and keep the out of fold predictions for the base learners, then fit the base learners on the whole dataset and fit the meta learners on the hold out predictions from the base learners.
Hi Jason !
I’m treating a seq2seq problem, in whish the output sequence has 5 components.
I builded, trainded and saved separatly 3 sub-models each one predict one component. The accuracy of each model alone is good but i want to have a good accuracy on the whole output sequence (5 sub-models toghether !! )
I did a simple concatenation (String concatenation) of the 5 predictions that the 5 sub-models gives to construct the whole sequence but this gives a bad accuracy in the whole output sequence !!
any suggestion ?? should i train the 5 sub-models toghether? how?
Knowing that the 5 sub-models have the same inputs.
thank you Jason for helping me !
You can train and save each model separately, use them to make predictions on a hold out set, then train another model to best combine their predictions.
It may or may not result in a lift in performance. Also try adding in the original input to the model that combines predictions.
Thank you Jason for this reply.
I don’t understand how to do this “use them to make predictions on a hold out set” ? what do you mean by hold out set ?
Thank you
Perhaps this will help:
https://machinelearningmastery.com/out-of-fold-predictions-in-machine-learning/
ok thank you Sir
You’re welcome.
Hi Jason,
Nice post.
Just one question, you trained and evaluated on the same dataset for the stacked model. Will that bias the performance metrics since the model already knows the true answer?
Yes, very likely.
https://machinelearningmastery.com/faq/single-faq/why-do-you-use-the-test-dataset-as-the-validation-dataset
Can we use np.hstack() directly instead of np.dstack()? so that can remove the reshape step.
You can concat arrays anyway you like!
I believe I am using dstack to ensure we are stacking arrays on the last dimension.
Hi jason,
I followed your procedure for my model but I get the error
ValueError: Error when checking model input: the list of Numpy arrays that you are passing to your model is not the size the model expected. Expected to see 2 array(s), but instead got the following list of 1 arrays:
I have both the X_train and X_val as list with two arrays of the same type. Do you know what could be wrong?
Perhaps start with the working example in the tutorial and slowly adapt it for your own models.
Hello,
first of all thank you for sharing your code, was really helpful.
I have a question concerning the input data for the integrated model. So as I understood is that the model takes 5 sets of input data , each set for each sub-model, let’s say i have sub-models that take the same set of input data, how can i manage to feed the model the same set of data without having to feed it 5 times? so mainly i want to feed the same set of input data for the 5 sub-models.
You’re welcome.
Load the data once and provide it to each model. Perhaps I don’t understand the problem?
I want to feed the model the same input data as the sub-models
I am serving the ensemble in tensorflow serving and with this code as is I am obliged to call the same field 5 times each time with the prefix ‘ensemble_i’. based on your reply, is there a way to implement a layer on top of this ensemble to be able to feed the data once so that my model takes this one set of data and replicate it 5 times then modify the name of the fields to become corresponding to the name of the layers (enseble_i_feature1, ensemble_2_feature2).
the 5 models take the same data as input. Or any other option?
Yes, you can wrap them in a single model, a so-called multi-input model.
We do exactly this in the section “Integrated Stacking Model”. Perhaps re-read that section?
Sorry, I don’t follow. Perhaps you can elaborate?
yes I am following the integrated model, but i would like to make it single input instead of multi input, a single input fed to the 5 sub-models
Perhaps write a for-loop?
Hi Jason , thank you very much for this tutorial. I have some questions, 1) how models can be visualized? 2) which kind of information can be extracted from models, and how? I want to compare two models to understand their similarities, correlation, …
Thanks again
Perhaps focus on comparing their predictive skill.
Hey Jason, thank you for this tutorial it was very helpful! I’m doing a project similar to the code you have used in your “Integrated Stacking Model” example, but with 3 separate fully connected networks (some with different input shapes, as it’s collecting different features on the same data) and I’m just caught on a bit of a snag with the error:
ValueError: The name “dense_1_input” is used 3 times in the model. All layer names should be unique.
I’ve noticed others encounter a similar problem, and was wondering if you could shed some light on the issue. Here is what I’ve already tried:
– using model.summary() before and after the layer name change to check it has successfully changed (it has)
– renaming all the names of the models themselves, as i noticed in model.summary() they were all named “sequential_1″
– using model.get_layer(name=’dense_1_input’).name=”NEWNAME” to manually change the input layer’s name for each model (instead raises the error ‘ValueError: No such layer: dense_1_input’)
– copying your completed block of code, only changing the code to load my 3 models instead of your 5
Everything I’ve tried raises that ‘dense_1_input’ error, even though the name is nowhere in the models!! The one place I saw it prop up is when I decided to run:
ensemble_visible = [print(model.input) for model in members].
Running that code returns:
Tensor(“dense_1_input:0”, shape=(None, 3), dtype=float32)
Tensor(“dense_1_input_1:0”, shape=(None, 1), dtype=float32)
Tensor(“dense_1_input_2:0”, shape=(None, 1), dtype=float32)
Note: this was after the layer names were changed.
Could this be the issue? Thanks again in advance!!
Perhaps try using a separate input layer for each model and set a unique name in the constructor of the layer.
Is there a reason the submodels are not trainable? Is that a limitation of Keras or is it intentional? Thanks
By design – we train them then figure out how to best combine them.
Hi Jason – what’s the difference between a weighted average ensemble and stacking with a linear model meta-learner (linear regression)? To me, it seems like they have similar mechanisms, where you find a linear combination of weights and base model outputs. I’m obviously missing something obvious!
Very little other than their name perhaps.
Hi, Jason!
Your tutorial is really helpful, but I can’t understand how I can combine two different types of neural networks as submodels(one of them CNN and another one Deep neural network),when each submodels have different input shape(for example (n_rows, 12) and (n_rows, 59)) and also they are training on different features, but for one classification goal with neural network as meta liner.
Thanks.
Sorry to hear that, not sure how I can help exactly. Perhaps try debugging the cause of the fault then address it?
Hey jason, really blown by the content and helped me miles for the work, i was able to interpret the ensemble, one question though, as i am applying this on a multilabel classification, while running the epocs it crashes and gives the error of logits and labels must be broadcastable: logits_size=[50,5] labels_size=[250,2] this 2 symbolizes that the label it is tkaing is 2 while i have5 classes ,what shall be the mistake.
Can it only work on binary classification
Thanks!
You may have to change the number of nodes in the output layer to match number of classes and change the loss function to categorical cross entropy.
hello,
really thanks for what you do really interesting!
I would like to know if is possible to adapt what you do for multiclass classification, for binary classification.
I mean, it’s enough to set:
model.add(Dense(2, activation=’sigmoid’))
And make centre=2 in generation of blobs?
Really thanks
Yes, this will help:
https://machinelearningmastery.com/faq/single-faq/how-can-i-change-a-neural-network-from-regression-to-classification
thanks for your quick answer!
The doubt was linked to the fact that I have a binary classification, I make two clusters, and of course in the output layer I need two neuron , otherwise it does not match.
But normally, don’t we have one neuron for binary classification , in output?
Thanks
A single node is used for output in a binary classification task.
Pardon, but when I insert 1 neuron in output, like this
def evaluate_model(X_train_, y_train_, X_validation_, y_validation_):
# define model
model = Sequential()
model.add(Dense(4, input_dim=2, activation=’relu’))
model.add(Dense(4, input_dim=2, activation=’relu’))
model.add(Dense(1, activation=’sigmoid’))
model.compile(loss=’binary_crossentropy’, optimizer=’adam’, metrics=[‘accuracy’])
# fit model
model.fit(X_train_, y_train_, epochs=100, verbose=0)
# evaluate the model
_, test_acc = model.evaluate(X_validation_, y_validation_, verbose=0)
return test_acc
# generate 2d classification dataset
X_new, y_new = make_blobs(n_samples=500, centers=2, n_features=2, cluster_std=2, random_state=2)
y_new = to_categorical(y_new)
I Have this error:
Error when checking target: expected dense_315 to have shape (1,) but got array with shape (2,
I don’t have it when I put 2 neuron, it seems really strange. I’m speaking always about binary classification.
It sounds like you have one hot encoded your binary label. You can change this back to a single variable with 0 and 1 values using argmax and then fit your model.
and of course change categorical, in binary_crossentropy
Hi Jason,
Thanks for your great tutorial
If we use validation set to train level 1 model, how can we tune the stacked model hyperparameters (such as number of hidden nodes)?
Perhaps cross-validate the whole thing.
Perhaps use a separate hold out dataset.
Can you tell me any research paper from where I can learn more about Integrated stacking ensemble?I can’t find such publication..
Perhaps try: scholar.google.com
Hi Jason,
Thank you for sharing the nice tutorial on stacking ensemble methods for Deep Learning.
One thing that bugged me is the way you have evaluated the accuracy of your total model. You trained the level 1 model on a certain Test Dataset and then calculated the accuracy on the same dataset.
How can you be certain that the increase in the accuracy you report is not due to overfitting on the test dataset?
Agreed, I should have used a hold out dataset:
https://machinelearningmastery.com/faq/single-faq/why-do-you-use-the-test-dataset-as-the-validation-dataset
Jason how to I plot multiple test sets on a given training set…ie I wannt 1 training loss curve and more then one test1 test2 test3.. curves on the same model.
You have one curve for each dataset, not each sample.
You can call the plot function many times with different data to get multiple lines on the same figure.
Hello Jason, Thanks for this tutorial. I used some of the code you presented in the section “integrated stacking model” to do some federated learning by stacking. Unfortunately I have an issue with the time it takes to train the meta-model which seems very long to me. I my setup, each partner trains a main model on its own data and I use a meta-model to aggregate the outputs of each partners’ model. It takes two to ten time longer to fit a small metamodel then to do the whole classical federated learning training, which is suprising to me. Indeed, the meta-model is rather small (400 wheights) compared to train a main model by federated learning. Do you have an idea on why it takes so long to train the integrated stacking model? and do you recommand something to fasten the code?
You’re welcome.
It may be slow because of the large number of weights.
For anyone who’s using the sample code and runs into the error of “AttributeError: can’t set attribute”. You can fix it by chanint it to
layer._name = ‘ensemble_’ + str(i + 1) + ‘_’ + layer.name
This is because there is an underlying hidden attribute _name, which causes the conflict.
PS: I have replaced all keras with tensorflow.keras
Ref: https://stackoverflow.com/a/56444135
Thanks for the note, what version of Keras are you using?
Hello, thank you so much for your article, it really helps me with research for my master thesis.
I have a little trouble with Raw Input Data extension. Should this be considered as an additional layer to the meta-model, or rather should the meta-model, after concatenating different models, be trained on the data again, using .fit()?
No, just more input features to the met a model.
Hello,
I have one question (propably i miss understand something)
Suppose:
We are changing meta-learner to XGBoost algorythm, and set max_depth= 100.
We got almost perfect fit. We fit model on stackX data, and test on stackX data. Why stackX data and testy labels aren’t split into additional test data? Predicting on train data isn’t good measure efficient of model
Ideally we would fit the meta model on the hold out predictions made by the base models (e.g. via cross-validation or a train/val split), then evaluate the whole system on a separate hold out dataset.
Hi, Jason. Thank you for the article. I have a bit confusing regarding the ensemble technique.
Suppose, the shape of data is (1316, 108164). For training purpose I have split the data into 3863. So, the dimension of each split is (1316, 28) and it creates 3863 autoencoder models. Each model is trained on (761, 28) samples and loss function is ‘mse’. I have trained and saved all the sub models.
By following Separate Stacking Model approcah, I have load all the saved models and evaluate each single model on the test dataset (1316, 28). The test dataset has 3863 splits. As there are 3863 models, I have evaluated each model with each split of data for example evaluate model_1 with split_1 test data, evaluate model_2 with split_2 data.
Now I want to evaluate the entire model from the sub-models. What is the best way to evaluate the loss of entire model?
That is a lot of models!
Ideally you want to evaluate the whole model on data not used to fit the base-models or the meta-model.
Thank you for the excellent blog. I test the code above and I have seen that fitting and testing on the same data with the stacked model is not appropriate. Because when I test it on unseen data the loss increases from .4 to .5.
# define ensemble model
stacked_model = define_stacked_model(members)
# fit stacked model on test dataset
fit_stacked_model(stacked_model, testX, testy)
# make predictions and evaluate
yhat = predict_stacked_model(stacked_model, testX)
yhat = argmax(yhat, axis=1)
acc = accuracy_score(testy, yhat)
print(‘Stacked Test Accuracy: %.3f’ % acc)
Yes, it is a good idea to test on a hold out dataset. I did not to keep the example simple.
Is it possible to stack sklearn model with a keras model or XGboost with a keras model?
Perhaps.
If you use the sklearn wrappers for each and the built-in stacking model:
https://machinelearningmastery.com/stacking-ensemble-machine-learning-with-python/
Hi Jason, thanks for your great tutorial! I’ve got a question about stacking models. Is it possible to stack LSTMs and XGBoost/RF models? 🙂
Yes, although not easily. It will require custom code.
Hi Jason, thanks for your amazing tutorial ! Have you post any topic or code related to fuzzy logic.
Sorry, I do not.
In the complete example of fitting a linear meta-learner for the stacking ensemble of MLP sub-models, line 71:
model = fit_stacked_model(members, testX, testy)
this one is supposed to fit training X, y. If we fit testing data and then predict testX, it will output the good result for sure.
Sorry, I don’t understand your question/comment, can you please rephrase or elaborate?
Hi Jason,
Thanks very much first of all for this tutorial, and in general for your work.
I have the same question, let me try to rephrase the question.
In line 80: fit_stacked_model(stacked_model, testX, testy), you are fitting the stacked model using the testing data, and then you are evaluating again using the same data.
Shouldn’t you use different data for training and evaluation?
Thanks,
Kostas
Yes, this is a common question that I answer here:
https://machinelearningmastery.com/faq/single-faq/why-do-you-use-the-test-dataset-as-the-validation-dataset
“The input layer for each of the sub-models will be used as a separate input head to this new model.”
I am trying to build a model to have an ensemble with only one head. Is there a possible solution?
Best regards,
Emanuele
Perhaps. You may have to experiment with the functional APi. Might get messy.
How can I plot loss and val_loss of the above-Stacked Generalization Ensemble of Neural Network Models?
thanks
It’s tricky because the models are trained on different data.
Nevertheless, perhaps you can save each trace to file and plot them at the end.
I use train test split to separate my data when training the models within the ensemble. Is it useful to train each model in the ensemble with a different random state? Would this allow me to capture the variance that may be lost when useful data is pulled into the test split instead of the train split, or would this overfit? Thanks for the great articles, these truly are a life saver!
Yes.
This might help with this line of thinking:
https://machinelearningmastery.com/different-results-each-time-in-machine-learning/
And this:
https://machinelearningmastery.com/evaluate-skill-deep-learning-models/
thank you for your valuable tutorials. I just wanted to know more about the reason why you used Logistic regression?
Thanks.
It’s common to use a linear model to combine predictions – it works well and is well understood.
I have folloed your tutorial for my time series univariant data .But the estimator gives n number of predictions based on n number of submodels used . But i am supposed to have one column to make evaluation . how can i solve this problem ?
Sorry, I don’t know the cause of your issue. Perhaps you can simplify your example to help expose the cause of the fault.
How do i ensemble cnn, rnn and lstm models
Same way as MLPs, no difference really, average the predictions together or use a model to average the predictions together.
I have a query about the model and that is , you have trained the level0 with the training dataset , after that you trained the level1 with test dataset/ validation dataset . the whole model is now trained with the training and validation dataset or you can say the model has seen the data’s. But if you do evaluation on the testx, will it be considered as generalized model ? the model also seem the testx , t]and corresponding groundtruth testy right ?
Yes, the model should not see the test dataset in anyway.
hi! great post – any suggestions/tips for using the Keras ImageDataGenerator instead of loading in x_train, y_train data?
Thanks.
Yes, perhaps start here:
https://machinelearningmastery.com/how-to-load-large-datasets-from-directories-for-deep-learning-with-keras/
Hey, great work!
I have an error: ValueError: The name “conv2d_input” is used 3 times in the model. All layer names should be unique. at tf.keras.models.Model(inputs=ensemble_visible, outputs=output).
I have 3 model (CNNs) that can predict 8 classes. Do you now what might be the cause?
Not off hand, perhaps you can post your code and error message to stackoverflow.com
Hello , thanks for your tutorial , it is such an amazing tutorial i have seen ever. I have a question :you used the the combination of level 0 predicted output and test data for training the meta learner . Then you evaluated the model using the same test data , will it be considered as generalized model ?
It is better to use a validation set to train level0, then use a separate test set to evaluate the complete model.
I use the same in both cases to keep the example simple:
https://machinelearningmastery.com/faq/single-faq/why-do-you-use-the-test-dataset-as-the-validation-dataset/
Thank you, Jason!
Have a nice day!
You’re welcome.
Thank you for the tutorial. I wanted to apply the same idea but with ImageDataGenerator, so I stacked the models that were trained using train batches created by a preprocessing_function but when I tried to fit the stacked model and feed it a train_generator, it gave me an error that there is no suitable input format with a list of ImageDataGenerator.
Do you have any solution or suggestion to make it works?
Thanks
You’re welcome.
Perhaps you can train each model, one at a time, save them, then train a level 1 model on the predictions made by each level 0 model.
Hi mr. Jason , thank you for your post it was very helpful.
i would like reuse your code for a multi class problem, i have some models trained on x ray image to get an 11 classe output , when i fit the staking model i get this errors :
y should be a 1d array, got an array of shape (1085, 11) instead.
Ensure your data matches the expectation of the model or the model matches the shape of your data.
I was following the “Integrated Stacking Model” part because my submodels are neural network-based models, so to train a level 1 model, I need the layers to be connected as you did in your example, the stacked model needs to have multiple inputs but following your comment, do I need a different logic for the stacked model ?
Thanks,
Sorry, I don’t understand your question. Perhaps you could rephrase or elaborate?
In the separate stacking model section, when evaluating standalone models on test dataset, why do we have to encode testy using to_categorical() ?
Because neural nets must work with numbers.
I understand. I was under the impression that we were using the “y” that was already converted using to_categorical(). But, I have later found out that, we are using the testy that is not already converted using to_categorical() and in order to comply with the saved models we are encoding the testy using to_categorical(). Thank you.
No problem.
Hi
I created 5 sub-models (different neural networks) using my database. Then, I created a Weighted Average Ensemble (WAE) model with a differential evolution algorithm using the 5 sub-models.
And I also created an Integrated Stacking Ensemble (ISE) using the 5 sub-models and a neural network with one hidden layer (as meta-learner).
The accuracy of ISE is 0.96 and The accuracy of WAE is 0.99.
I can not understand why ISE is worse than WAE? I think a linear combination with values would provide the same results. Can you help me to realize this issue? Why meta leaner in ISE can not find a good linear combination. Maybe the selected nonlinear activation function for the additional NN makes things worse. Am I missing something?
I cannot explain accurately but one thing for sure is that ISE is not linear. It is more like nested functions that further process from previous step. Some data would be more suitable to use linear model. If that’s your case, may be ISE is overfitting.
Hi Json,
May I know why you used test set data for training of the final model, why not training set (originally used to train sub models) ?
As the final model layers (trainable layers) havent seen the training data, we should ideally train on training set right?
I think so. Probably that’s a typo.
Hello,
Thank you so much for this work, it is well-explained. I tried to implement your stacked method to combine (CNN, LSTM, BiLSTM, and GRU) using the following architecture :
def CNN(input_shape):
X_indices = Input(input_shape)
embeddings = embedding_layer(X_indices)
X = Conv1D(512, 3, activation=’relu’)(embeddings)
X = MaxPooling1D(3)(X)
# X = Conv1D(256, 3, activation=’relu’)(X)
# X = MaxPooling1D(3)(X)
X = Conv1D(256, 3, activation=’relu’)(X)
X = Dropout(0.8)(X)
X = MaxPooling1D(3)(X)
X = GlobalMaxPooling1D()(X)
X = Dense(256, activation=’relu’)(X)
X = Dense(1, activation=’sigmoid’)(X)
model = Model(inputs=X_indices, outputs=X)
return model
def LSTM_(input_shape):
X_indices = Input(input_shape)
embeddings = embedding_layer(X_indices)
X = LSTM(128, return_sequences=True)(embeddings)
X = Dropout(0.6)(X)
# X = LSTM(128, return_sequences=True)(X)
# X = Dropout(0.6)(X)
X = LSTM(128)(X)
X = Dense(1, activation=’sigmoid’)(X)
model = Model(inputs=X_indices, outputs=X)
return model
def BiLSTM(input_shape):
X_indices = Input(input_shape)
embeddings = embedding_layer(X_indices)
X = Bidirectional(LSTM(128, return_sequences=True))(embeddings)
X = Dropout(0.6)(X)
X = Bidirectional(LSTM(128, return_sequences=True))(X)
X = Dense(1, activation=’sigmoid’)(X)
model = Model(inputs=X_indices, outputs=X)
return model
def GRU_(input_shape):
X_indices = Input(input_shape)
embeddings = embedding_layer(X_indices)
X = GRU(128, return_sequences=True)(embeddings)
X = Dropout(0.6)(X)
X = GRU(128, return_sequences=True)(X)
X = Dropout(0.6)(X)
X = Dense(1, activation=’sigmoid’)(X)
model = Model(inputs=X_indices, outputs=X)
return model
maxLen = 150
X_train_indices = tokenizer.texts_to_sequences(X_train)
X_train_indices = pad_sequences(X_train_indices, maxlen=maxLen, padding=’post’)
adam = keras.optimizers.Adam(learning_rate=0.0001)
model1 = CNN((maxLen,))
print(model1.summary())
model1.compile(optimizer=’adam’, loss=’binary_crossentropy’, metrics=[‘accuracy’])
model1.fit(X_train_indices, Y_train, batch_size=64, epochs=1)
model1.save(‘model1.h5′)
print(“model 1 CNN saved”)
model2 = LSTM_((maxLen,))
print(model2.summary())
model2.compile(optimizer=’adam’, loss=’binary_crossentropy’, metrics=[‘accuracy’])
model2.fit(X_train_indices, Y_train, batch_size=64, epochs=1)
model2.save(‘model2.h5′)
print(“model 2 LSTM saved”)
model3 = BiLSTM((maxLen,))
print(model3.summary())
model3.compile(optimizer=’adam’, loss=’binary_crossentropy’, metrics=[‘accuracy’])
model3.fit(X_train_indices, Y_train, batch_size=64, epochs=1)
model3.save(‘model3.h5′)
print(“model 3 BiLSTM saved”)
model4 = GRU_((maxLen,))
print(model4.summary())
model4.compile(optimizer=’adam’, loss=’binary_crossentropy’, metrics=[‘accuracy’])
model4.fit(X_train_indices, Y_train, batch_size=64, epochs=1)
model4.save(‘model4.h5’)
print(“model 4 GRU saved”)
It works when I combine two models, but when I try to combine the 4 models it gives me this error :
all the input array dimensions for the concatenation axis must match exactly, but along dimension 1, the array at index 0 has size 1 and the array at index 1 has size 1
Can you help me to solve this problem. Thank you.
Hi Nassera…What line in your code is identified as having the error?
Thank you for your reply, the error on stacked_dataset function (in reshape function) Note that, I implemented your code on twitter dataset.
I inserted some ‘print’ into the function to display the different dimensions as follows :
def stacked_dataset(members, inputX):
stackX = None
for model in members:
# make prediction
yhat = model.predict(inputX)
print(“yhat”, yhat.shape)
# y = np.argmax(yhat, axis=1) , verbose=0
# stack predictions into [rows, members, probabilities]
if stackX is None:
stackX = yhat #
else:
stackX = np.vstack((stackX, yhat))
print(‘stackX’, stackX.shape)
# flatten predictions to [rows, members x probabilities]
stackX = stackX.reshape((stackX.shape[0], (stackX.shape[1]*stackX.shape[2])))
print(‘stackX after reshape’, stackX.shape)
return stackX
Igot this error :
model 4 GRU saved
>loaded models
Loaded the models
yhat (39891, 1)
yhat (39891, 1)
stackX (79782, 1)
yhat (39891, 150, 1)
Traceback (most recent call last):
File “C:/Users/admin/PycharmProjects/MyProject/Ensemble DL/MDSA.py”, line 229, in
modelStacked = fit_stacked_model(members, X_train_indices, Y_train)
File “C:/Users/admin/PycharmProjects/MyProject/Ensemble DL/MDSA.py”, line 220, in fit_stacked_model
stackedX = stacked_dataset(members, inputX)
File “C:/Users/admin/PycharmProjects/MyProject/Ensemble DL/MDSA.py”, line 210, in stacked_dataset
stackX = np.vstack((stackX, yhat))
File “”, line 5, in vstack
File “C:\Users\admin\PycharmProjects\MyProject\venv\lib\site-packages\numpy\core\shape_base.py”, line 283, in vstack
return _nx.concatenate(arrs, 0)
File “”, line 5, in concatenate
ValueError: all the input arrays must have same number of dimensions, but the array at index 0 has 2 dimension(s) and the array at index 1 has 3 dimension(s)
Hello,
I have 4 models, and their output shapes are as follows:
predict CNN model (9973, 1)
predict LSTM model (9973, 1)
predict BiLSTM model (9973, 150, 1)
predict GRU model (9973, 150, 1)
How can I stack and pass those outputs to Logitic regression?
Thank you.
hey!!!
can u tell how may I stacked 3 models having different input shape arrays??????
as i’m using DNN (2D array), CNN(3D array), LSTM(3D array)
Hello Halima…The following should help add clarity:
https://www.geeksforgeeks.org/stacking-in-machine-learning-2/
Hello Jason this is a great tutorial. I’m just trying to better understand how (or if) it can fits my use case.
I have several datasets containing the same features set, each. They belong to a group of similar operating machines, whose operations is represented by those features. One dataset for each machine.
We currently have many regressions models based on LSTM Autoencoder, one model for each of the machines. This works quite well in modelling the features.
Now I’m working on similar model that should be capable to learn from all the datasets and generalize by a single model. It should take as input the features set, regardless it belongs to one machine or the other.
Is stacking ensemble the right way ? Any advice for that ? Really much appreciated ! Thanks a lot
Hi Mike…the following resource may be of interest to you:
https://machinelearningmastery.com/ensemble-machine-learning-with-python-7-day-mini-course/
Your meta learner is cheating, it is using test_y which in practice would normally be on kaggle servers so you don’t have access to it. if you try to use train data for the meta learner then the submodels will be asked to predict points that they have already seen, so the meta learner will perform really poorly. you could try having 3 different data sets so one is used to train the submodels, one for the meta learner, and the last one is used as the test set, but then the submodels have less data to work with and perform worse.
Thank you for the feedback Timothe!
Hi James,
I have manually created 5 sub NN models, and then applied your example to loading the models as defined and then tried to adapt it to LinearRegression.
I was succesful with the initial model, using a LinearRegressor as the meta-learner.
yhat = stacked_prediction(members, model, X_val)
mae = mean_absolute_error(y_val, yhat)
print(‘Stacked Mean Abs Error: %.3f’ % mae)
#0.094
So, yeah, using mean_absolute_error as my estimate.
However, when adapting the example with a neural network as a meta-learner i get:
fit_stacked_model(stacked_model, X_train, y_train)
# make predictions and evaluate
yhat = predict_stacked_model(stacked_model, X_val)
mae = mean_absolute_error(y_val, yhat)
print(‘Stacked Mean abs Error: %.3f’ % mae)
#mae = 2.1
In addition, changing verbose = 1, i can see that my model does not really improve from training after a few epochs:
Epoch 1/300
1000/1000 [==============================] – 7s 5ms/step – loss: 0.3456 – mae: 0.4919
Epoch 2/300
1000/1000 [==============================] – 4s 4ms/step – loss: 0.2513 – mae: 0.4444
Epoch 3/300
1000/1000 [==============================] – 4s 4ms/step – loss: 0.2513 – mae: 0.4444
Epoch 4/300
1000/1000 [==============================] – 4s 4ms/step – loss: 0.2512 – mae: 0.4444
Epoch 5/300
1000/1000 [==============================] – 4s 4ms/step – loss: 0.2512 – mae: 0.4444
..
..
Epoch 41/300
1000/1000 [==============================] – 5s 5ms/step – loss: 0.2506 – mae: 0.4444
Epoch 42/300
1000/1000 [==============================] – 5s 5ms/step – loss: 0.2506 – mae: 0.4444
Epoch 43/300
1000/1000 [==============================] – 5s 5ms/step – loss: 0.2506 – mae: 0.4444
Epoch 44/300
772/1000 [======================>…….] – ETA: 1s – loss: 0.2506 – mae: 0.4444
I’m not sure why it does that?
Thank you for your time.
Hi Ian…You may find the following resource helpful:
https://machinelearningmastery.com/learning-curves-for-diagnosing-machine-learning-model-performance/
great post
I have a question
can we use this code for binary image classification?
please sir I am waiting for your response.
Hi kamranullah…you are very welcome! The example is a great starting point along with the following resource:
https://machinelearningmastery.com/binary-classification-tutorial-with-the-keras-deep-learning-library/
I am using ImageDatagenerator class to generate dataset on the fly so in place of testy i am using validation_set.class_indices.
and it is throughing an error. plz help
Thank you for your detailed explanation.
There is something that I do not understand:
in the section “Separate Stacking Model”
you created a stacking model from 5 sub-models where each of them has 3 predictions for each class
Then you updated the dataset for the stacking model by changing the size of the test to [1000,15].
In the next step you choose logistic regression for the classifier of the stacking model, but now I am confused.
Even if we say that we can use logistic regression for Multi-Class Classification by the method of “one vs rest”
by taking the argmax from all binary classifications, it will be fine if we have 3 classes but now we have 15!
so what is the meaning of choosing the argmax of class “i” (where I>3)?
Hi DBM…The following resource provides another view that may be of interest:
https://www.analyticsvidhya.com/blog/2021/08/ensemble-stacking-for-machine-learning-and-deep-learning/
If the predicted class probabilities are correct and equal to 1.0000 for each base model, but in the ensemble model, the predicted class probabilities for the correct class decrease, for example, to 0.9821, and there are probabilities assigned to other classes, like 0.179. Is there any specific reason for the result I obtained?
Hi Firly…The following resource may be of interest regarding best practices for when to use ensemble learning. It may not always be an advantage.
https://machinelearningmastery.com/why-use-ensemble-learning/
Thank you for the answer my question, can I ask you something I’m working on a manual calculation case study about integrated ensemble stacking, May I know how the process proceeds after the concatenation process from the 2 models into the hidden layer? Does it consist of a 2-dimensional matrix? Isn’t it in the hidden layer that only 1-dimensional matrices should enter?