Recursive Feature Elimination, or RFE for short, is a popular feature selection algorithm.
RFE is popular because it is easy to configure and use and because it is effective at selecting those features (columns) in a training dataset that are more or most relevant in predicting the target variable.
There are two important configuration options when using RFE: the choice in the number of features to select and the choice of the algorithm used to help choose features. Both of these hyperparameters can be explored, although the performance of the method is not strongly dependent on these hyperparameters being configured well.
In this tutorial, you will discover how to use Recursive Feature Elimination (RFE) for feature selection in Python.
After completing this tutorial, you will know:
- RFE is an efficient approach for eliminating features from a training dataset for feature selection.
- How to use RFE for feature selection for classification and regression predictive modeling problems.
- How to explore the number of selected features and wrapped algorithm used by the RFE procedure.
Kick-start your project with my new book Data Preparation for Machine Learning, including step-by-step tutorials and the Python source code files for all examples.
Let’s get started.

Recursive Feature Elimination (RFE) for Feature Selection in Python
Taken by djandywdotcom, some rights reserved.
Tutorial Overview
This tutorial is divided into three parts; they are:
- Recursive Feature Elimination
- RFE With scikit-learn
- RFE for Classification
- RFE for Regression
- RFE Hyperparameters
- Explore Number of Features
- Automatically Select the Number of Features
- Which Features Were Selected
- Explore Base Algorithm
Recursive Feature Elimination
Recursive Feature Elimination, or RFE for short, is a feature selection algorithm.
A machine learning dataset for classification or regression is comprised of rows and columns, like an excel spreadsheet. Rows are often referred to as samples and columns are referred to as features, e.g. features of an observation in a problem domain.
Feature selection refers to techniques that select a subset of the most relevant features (columns) for a dataset. Fewer features can allow machine learning algorithms to run more efficiently (less space or time complexity) and be more effective. Some machine learning algorithms can be misled by irrelevant input features, resulting in worse predictive performance.
For more on feature selection generally, see the tutorial:
RFE is a wrapper-type feature selection algorithm. This means that a different machine learning algorithm is given and used in the core of the method, is wrapped by RFE, and used to help select features. This is in contrast to filter-based feature selections that score each feature and select those features with the largest (or smallest) score.
Technically, RFE is a wrapper-style feature selection algorithm that also uses filter-based feature selection internally.
RFE works by searching for a subset of features by starting with all features in the training dataset and successfully removing features until the desired number remains.
This is achieved by fitting the given machine learning algorithm used in the core of the model, ranking features by importance, discarding the least important features, and re-fitting the model. This process is repeated until a specified number of features remains.
When the full model is created, a measure of variable importance is computed that ranks the predictors from most important to least. […] At each stage of the search, the least important predictors are iteratively eliminated prior to rebuilding the model.
— Pages 494-495, Applied Predictive Modeling, 2013.
Features are scored either using the provided machine learning model (e.g. some algorithms like decision trees offer importance scores) or by using a statistical method.
The importance calculations can be model based (e.g., the random forest importance criterion) or using a more general approach that is independent of the full model.
— Page 494, Applied Predictive Modeling, 2013.
Now that we are familiar with the RFE procedure, let’s review how we can use it in our projects.
Want to Get Started With Data Preparation?
Take my free 7-day email crash course now (with sample code).
Click to sign-up and also get a free PDF Ebook version of the course.
RFE With scikit-learn
RFE can be implemented from scratch, although it can be challenging for beginners.
The scikit-learn Python machine learning library provides an implementation of RFE for machine learning.
It is available in modern versions of the library.
First, confirm that you are using a modern version of the library by running the following script:
|
1 2 3 |
# check scikit-learn version import sklearn print(sklearn.__version__) |
Running the script will print your version of scikit-learn.
Your version should be the same or higher. If not, you must upgrade your version of the scikit-learn library.
|
1 |
0.22.1 |
The RFE method is available via the RFE class in scikit-learn.
RFE is a transform. To use it, first the class is configured with the chosen algorithm specified via the “estimator” argument and the number of features to select via the “n_features_to_select” argument.
The algorithm must provide a way to calculate important scores, such as a decision tree. The algorithm used in RFE does not have to be the algorithm that is fit on the selected features; different algorithms can be used.
Once configured, the class must be fit on a training dataset to select the features by calling the fit() function. After the class is fit, the choice of input variables can be seen via the “support_” attribute that provides a True or False for each input variable.
It can then be applied to the training and test datasets by calling the transform() function.
|
1 2 3 4 5 6 7 |
... # define the method rfe = RFE(estimator=DecisionTreeClassifier(), n_features_to_select=3) # fit the model rfe.fit(X, y) # transform the data X, y = rfe.transform(X, y) |
It is common to use k-fold cross-validation to evaluate a machine learning algorithm on a dataset. When using cross-validation, it is good practice to perform data transforms like RFE as part of a Pipeline to avoid data leakage.
Now that we are familiar with the RFE API, let’s take a look at how to develop a RFE for both classification and regression.
RFE for Classification
In this section, we will look at using RFE for a classification problem.
First, we can use the make_classification() function to create a synthetic binary classification problem with 1,000 examples and 10 input features, five of which are important and five of which are redundant.
The complete example is listed below.
|
1 2 3 4 5 6 |
# test classification dataset from sklearn.datasets import make_classification # define dataset X, y = make_classification(n_samples=1000, n_features=10, n_informative=5, n_redundant=5, random_state=1) # summarize the dataset print(X.shape, y.shape) |
Running the example creates the dataset and summarizes the shape of the input and output components.
|
1 |
(1000, 10) (1000,) |
Next, we can evaluate an RFE feature selection algorithm on this dataset. We will use a DecisionTreeClassifier to choose features and set the number of features to five. We will then fit a new DecisionTreeClassifier model on the selected features.
We will evaluate the model using repeated stratified k-fold cross-validation, with three repeats and 10 folds. We will report the mean and standard deviation of the accuracy of the model across all repeats and folds.
The complete example is listed below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 |
# evaluate RFE for classification from numpy import mean from numpy import std from sklearn.datasets import make_classification from sklearn.model_selection import cross_val_score from sklearn.model_selection import RepeatedStratifiedKFold from sklearn.feature_selection import RFE from sklearn.tree import DecisionTreeClassifier from sklearn.pipeline import Pipeline # define dataset X, y = make_classification(n_samples=1000, n_features=10, n_informative=5, n_redundant=5, random_state=1) # create pipeline rfe = RFE(estimator=DecisionTreeClassifier(), n_features_to_select=5) model = DecisionTreeClassifier() pipeline = Pipeline(steps=[('s',rfe),('m',model)]) # evaluate model cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1) n_scores = cross_val_score(pipeline, X, y, scoring='accuracy', cv=cv, n_jobs=-1, error_score='raise') # report performance print('Accuracy: %.3f (%.3f)' % (mean(n_scores), std(n_scores))) |
Running the example reports the mean and standard deviation accuracy of the model.
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
In this case, we can see the RFE that uses a decision tree and selects five features and then fits a decision tree on the selected features achieves a classification accuracy of about 88.6 percent.
|
1 |
Accuracy: 0.886 (0.030) |
We can also use the RFE model pipeline as a final model and make predictions for classification.
First, the RFE and model are fit on all available data, then the predict() function can be called to make predictions on new data.
The example below demonstrates this on our binary classification dataset.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 |
# make a prediction with an RFE pipeline from numpy import mean from numpy import std from sklearn.datasets import make_classification from sklearn.feature_selection import RFE from sklearn.tree import DecisionTreeClassifier from sklearn.pipeline import Pipeline # define dataset X, y = make_classification(n_samples=1000, n_features=10, n_informative=5, n_redundant=5, random_state=1) # create pipeline rfe = RFE(estimator=DecisionTreeClassifier(), n_features_to_select=5) model = DecisionTreeClassifier() pipeline = Pipeline(steps=[('s',rfe),('m',model)]) # fit the model on all available data pipeline.fit(X, y) # make a prediction for one example data = [[2.56999479,-0.13019997,3.16075093,-4.35936352,-1.61271951,-1.39352057,-2.48924933,-1.93094078,3.26130366,2.05692145]] yhat = pipeline.predict(data) print('Predicted Class: %d' % (yhat)) |
Running the example fits the RFE pipeline on the entire dataset and is then used to make a prediction on a new row of data, as we might when using the model in an application.
|
1 |
Predicted Class: 1 |
Now that we are familiar with using RFE for classification, let’s look at the API for regression.
RFE for Regression
In this section, we will look at using RFE for a regression problem.
First, we can use the make_regression() function to create a synthetic regression problem with 1,000 examples and 10 input features, five of which are important and five of which are redundant.
The complete example is listed below.
|
1 2 3 4 5 6 |
# test regression dataset from sklearn.datasets import make_regression # define dataset X, y = make_regression(n_samples=1000, n_features=10, n_informative=5, random_state=1) # summarize the dataset print(X.shape, y.shape) |
Running the example creates the dataset and summarizes the shape of the input and output components.
|
1 |
(1000, 10) (1000,) |
Next, we can evaluate an REFE algorithm on this dataset.
As we did with the last section, we will evaluate the pipeline with a decision tree using repeated k-fold cross-validation, with three repeats and 10 folds.
We will report the mean absolute error (MAE) of the model across all repeats and folds. The scikit-learn library makes the MAE negative so that it is maximized instead of minimized. This means that larger negative MAE are better and a perfect model has a MAE of 0.
The complete example is listed below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 |
# evaluate RFE for regression from numpy import mean from numpy import std from sklearn.datasets import make_regression from sklearn.model_selection import cross_val_score from sklearn.model_selection import RepeatedKFold from sklearn.feature_selection import RFE from sklearn.tree import DecisionTreeRegressor from sklearn.pipeline import Pipeline # define dataset X, y = make_regression(n_samples=1000, n_features=10, n_informative=5, random_state=1) # create pipeline rfe = RFE(estimator=DecisionTreeRegressor(), n_features_to_select=5) model = DecisionTreeRegressor() pipeline = Pipeline(steps=[('s',rfe),('m',model)]) # evaluate model cv = RepeatedKFold(n_splits=10, n_repeats=3, random_state=1) n_scores = cross_val_score(pipeline, X, y, scoring='neg_mean_absolute_error', cv=cv, n_jobs=-1, error_score='raise') # report performance print('MAE: %.3f (%.3f)' % (mean(n_scores), std(n_scores))) |
Running the example reports the mean and standard deviation accuracy of the model.
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
In this case, we can see the RFE pipeline with a decision tree model achieves a MAE of about 26.
|
1 |
MAE: -26.853 (2.696) |
We can also use the c as a final model and make predictions for regression.
First, the Pipeline is fit on all available data, then the predict() function can be called to make predictions on new data.
The example below demonstrates this on our regression dataset.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 |
# make a regression prediction with an RFE pipeline from numpy import mean from numpy import std from sklearn.datasets import make_regression from sklearn.feature_selection import RFE from sklearn.tree import DecisionTreeRegressor from sklearn.pipeline import Pipeline # define dataset X, y = make_regression(n_samples=1000, n_features=10, n_informative=5, random_state=1) # create pipeline rfe = RFE(estimator=DecisionTreeRegressor(), n_features_to_select=5) model = DecisionTreeRegressor() pipeline = Pipeline(steps=[('s',rfe),('m',model)]) # fit the model on all available data pipeline.fit(X, y) # make a prediction for one example data = [[-2.02220122,0.31563495,0.82797464,-0.30620401,0.16003707,-1.44411381,0.87616892,-0.50446586,0.23009474,0.76201118]] yhat = pipeline.predict(data) print('Predicted: %.3f' % (yhat)) |
Running the example fits the RFE pipeline on the entire dataset and is then used to make a prediction on a new row of data, as we might when using the model in an application.
|
1 |
Predicted: -84.288 |
Now that we are familiar with using the scikit-learn API to evaluate and use RFE for feature selection, let’s look at configuring the model.
RFE Hyperparameters
In this section, we will take a closer look at some of the hyperparameters you should consider tuning for the RFE method for feature selection and their effect on model performance.
Explore Number of Features
An important hyperparameter for the RFE algorithm is the number of features to select.
In the previous section, we used an arbitrary number of selected features, five, which matches the number of informative features in the synthetic dataset. In practice, we cannot know the best number of features to select with RFE; instead, it is good practice to test different values.
The example below demonstrates selecting different numbers of features from 2 to 10 on the synthetic binary classification dataset.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 |
# explore the number of selected features for RFE from numpy import mean from numpy import std from sklearn.datasets import make_classification from sklearn.model_selection import cross_val_score from sklearn.model_selection import RepeatedStratifiedKFold from sklearn.feature_selection import RFE from sklearn.tree import DecisionTreeClassifier from sklearn.pipeline import Pipeline from matplotlib import pyplot # get the dataset def get_dataset(): X, y = make_classification(n_samples=1000, n_features=10, n_informative=5, n_redundant=5, random_state=1) return X, y # get a list of models to evaluate def get_models(): models = dict() for i in range(2, 10): rfe = RFE(estimator=DecisionTreeClassifier(), n_features_to_select=i) model = DecisionTreeClassifier() models[str(i)] = Pipeline(steps=[('s',rfe),('m',model)]) return models # evaluate a give model using cross-validation def evaluate_model(model, X, y): cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1) scores = cross_val_score(model, X, y, scoring='accuracy', cv=cv, n_jobs=-1, error_score='raise') return scores # define dataset X, y = get_dataset() # get the models to evaluate models = get_models() # evaluate the models and store results results, names = list(), list() for name, model in models.items(): scores = evaluate_model(model, X, y) results.append(scores) names.append(name) print('>%s %.3f (%.3f)' % (name, mean(scores), std(scores))) # plot model performance for comparison pyplot.boxplot(results, labels=names, showmeans=True) pyplot.show() |
Running the example first reports the mean accuracy for each configured number of input features.
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
In this case, we can see that performance improves as the number of features increase and perhaps peaks around 4-to-7 as we might expect, given that only five features are relevant to the target variable.
|
1 2 3 4 5 6 7 8 |
>2 0.715 (0.044) >3 0.825 (0.031) >4 0.876 (0.033) >5 0.887 (0.030) >6 0.890 (0.031) >7 0.888 (0.025) >8 0.885 (0.028) >9 0.884 (0.025) |
A box and whisker plot is created for the distribution of accuracy scores for each configured number of features.
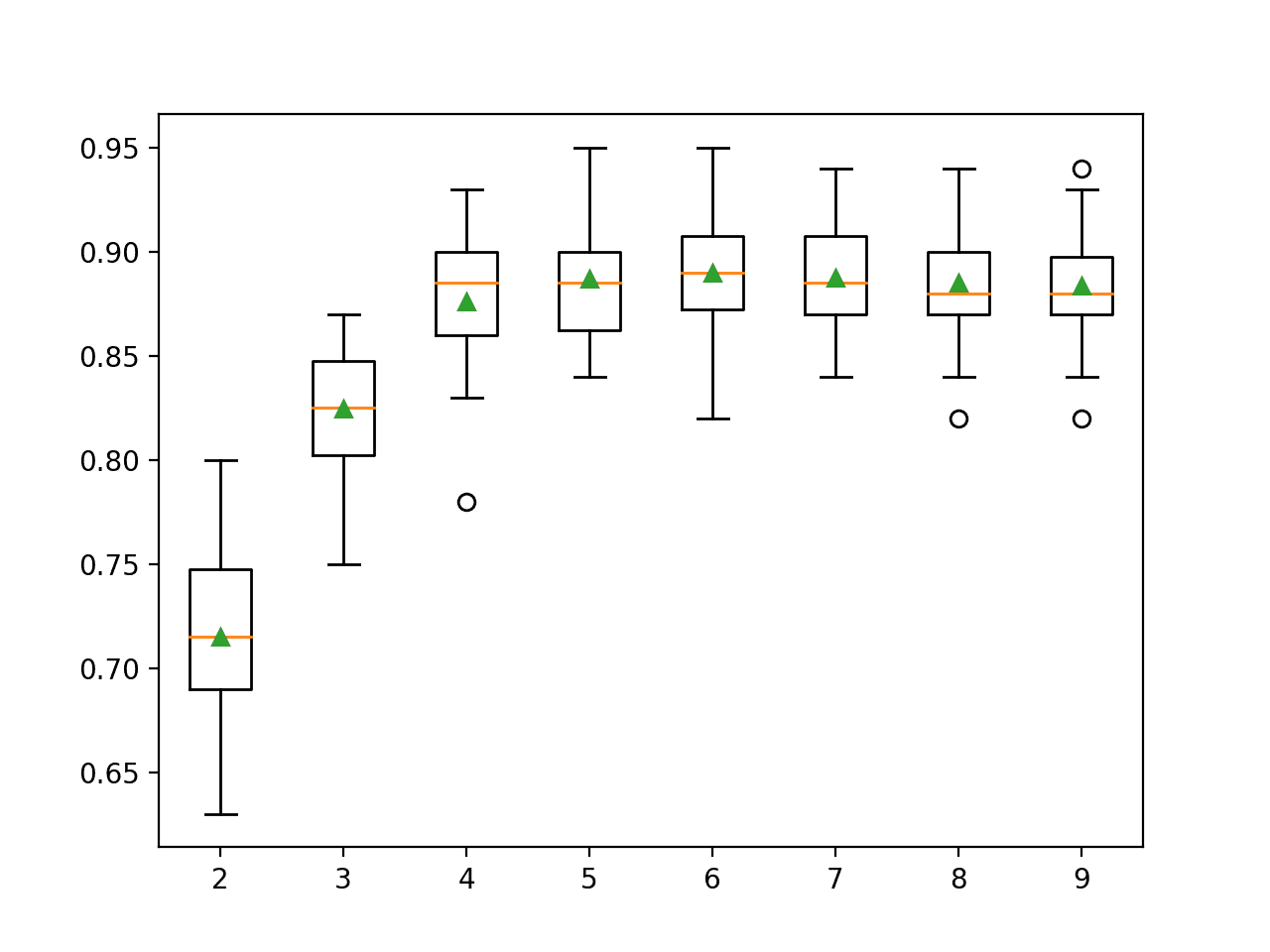
Box Plot of RFE Number of Selected Features vs. Classification Accuracy
Automatically Select the Number of Features
It is also possible to automatically select the number of features chosen by RFE.
This can be achieved by performing cross-validation evaluation of different numbers of features as we did in the previous section and automatically selecting the number of features that resulted in the best mean score.
The RFECV class implements this for us.
The RFECV is configured just like the RFE class regarding the choice of the algorithm that is wrapped. Additionally, the minimum number of features to be considered can be specified via the “min_features_to_select” argument (defaults to 1) and we can also specify the type of cross-validation and scoring to use via the “cv” (defaults to 5) and “scoring” arguments (uses accuracy for classification).
|
1 2 3 |
... # automatically choose the number of features rfe = RFECV(estimator=DecisionTreeClassifier()) |
We can demonstrate this on our synthetic binary classification problem and use RFECV in our pipeline instead of RFE to automatically choose the number of selected features.
The complete example is listed below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 |
# automatically select the number of features for RFE from numpy import mean from numpy import std from sklearn.datasets import make_classification from sklearn.model_selection import cross_val_score from sklearn.model_selection import RepeatedStratifiedKFold from sklearn.feature_selection import RFECV from sklearn.tree import DecisionTreeClassifier from sklearn.pipeline import Pipeline # define dataset X, y = make_classification(n_samples=1000, n_features=10, n_informative=5, n_redundant=5, random_state=1) # create pipeline rfe = RFECV(estimator=DecisionTreeClassifier()) model = DecisionTreeClassifier() pipeline = Pipeline(steps=[('s',rfe),('m',model)]) # evaluate model cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1) n_scores = cross_val_score(pipeline, X, y, scoring='accuracy', cv=cv, n_jobs=-1, error_score='raise') # report performance print('Accuracy: %.3f (%.3f)' % (mean(n_scores), std(n_scores))) |
Running the example reports the mean and standard deviation accuracy of the model.
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
In this case, we can see the RFE that uses a decision tree and automatically selects a number of features and then fits a decision tree on the selected features achieves a classification accuracy of about 88.6 percent.
|
1 |
Accuracy: 0.886 (0.026) |
Which Features Were Selected
When using RFE, we may be interested to know which features were selected and which were removed.
This can be achieved by reviewing the attributes of the fit RFE object (or fit RFECV object). The “support_” attribute reports true or false as to which features in order of column index were included and the “ranking_” attribute reports the relative ranking of features in the same order.
The example below fits an RFE model on the whole dataset and selects five features, then reports each feature column index (0 to 9), whether it was selected or not (True or False), and the relative feature ranking.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 |
# report which features were selected by RFE from sklearn.datasets import make_classification from sklearn.feature_selection import RFE from sklearn.tree import DecisionTreeClassifier # define dataset X, y = make_classification(n_samples=1000, n_features=10, n_informative=5, n_redundant=5, random_state=1) # define RFE rfe = RFE(estimator=DecisionTreeClassifier(), n_features_to_select=5) # fit RFE rfe.fit(X, y) # summarize all features for i in range(X.shape[1]): print('Column: %d, Selected %s, Rank: %.3f' % (i, rfe.support_[i], rfe.ranking_[i])) |
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
Running the example lists of the 10 input features and whether or not they were selected as well as their relative ranking of importance.
|
1 2 3 4 5 6 7 8 9 10 |
Column: 0, Selected False, Rank: 5.000 Column: 1, Selected False, Rank: 4.000 Column: 2, Selected True, Rank: 1.000 Column: 3, Selected True, Rank: 1.000 Column: 4, Selected True, Rank: 1.000 Column: 5, Selected False, Rank: 6.000 Column: 6, Selected True, Rank: 1.000 Column: 7, Selected False, Rank: 3.000 Column: 8, Selected True, Rank: 1.000 Column: 9, Selected False, Rank: 2.000 |
Explore Base Algorithm
There are many algorithms that can be used in the core RFE, as long as they provide some indication of variable importance.
Most decision tree algorithms are likely to report the same general trends in feature importance, but this is not guaranteed. It might be helpful to explore the use of different algorithms wrapped by RFE.
The example below demonstrates how you might explore this configuration option.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 |
# explore the algorithm wrapped by RFE from numpy import mean from numpy import std from sklearn.datasets import make_classification from sklearn.model_selection import cross_val_score from sklearn.model_selection import RepeatedStratifiedKFold from sklearn.feature_selection import RFE from sklearn.linear_model import LogisticRegression from sklearn.linear_model import Perceptron from sklearn.tree import DecisionTreeClassifier from sklearn.ensemble import RandomForestClassifier from sklearn.ensemble import GradientBoostingClassifier from sklearn.pipeline import Pipeline from matplotlib import pyplot # get the dataset def get_dataset(): X, y = make_classification(n_samples=1000, n_features=10, n_informative=5, n_redundant=5, random_state=1) return X, y # get a list of models to evaluate def get_models(): models = dict() # lr rfe = RFE(estimator=LogisticRegression(), n_features_to_select=5) model = DecisionTreeClassifier() models['lr'] = Pipeline(steps=[('s',rfe),('m',model)]) # perceptron rfe = RFE(estimator=Perceptron(), n_features_to_select=5) model = DecisionTreeClassifier() models['per'] = Pipeline(steps=[('s',rfe),('m',model)]) # cart rfe = RFE(estimator=DecisionTreeClassifier(), n_features_to_select=5) model = DecisionTreeClassifier() models['cart'] = Pipeline(steps=[('s',rfe),('m',model)]) # rf rfe = RFE(estimator=RandomForestClassifier(), n_features_to_select=5) model = DecisionTreeClassifier() models['rf'] = Pipeline(steps=[('s',rfe),('m',model)]) # gbm rfe = RFE(estimator=GradientBoostingClassifier(), n_features_to_select=5) model = DecisionTreeClassifier() models['gbm'] = Pipeline(steps=[('s',rfe),('m',model)]) return models # evaluate a give model using cross-validation def evaluate_model(model, X, y): cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1) scores = cross_val_score(model, X, y, scoring='accuracy', cv=cv, n_jobs=-1) return scores # define dataset X, y = get_dataset() # get the models to evaluate models = get_models() # evaluate the models and store results results, names = list(), list() for name, model in models.items(): scores = evaluate_model(model, X, y) results.append(scores) names.append(name) print('>%s %.3f (%.3f)' % (name, mean(scores), std(scores))) # plot model performance for comparison pyplot.boxplot(results, labels=names, showmeans=True) pyplot.show() |
Running the example first reports the mean accuracy for each wrapped algorithm.
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
In this case, the results suggest that linear algorithms like logistic regression might select better features more reliably than the chosen decision tree and ensemble of decision tree algorithms.
|
1 2 3 4 5 |
>lr 0.893 (0.030) >per 0.843 (0.040) >cart 0.887 (0.033) >rf 0.858 (0.038) >gbm 0.891 (0.030) |
A box and whisker plot is created for the distribution of accuracy scores for each configured wrapped algorithm.
We can see the general trend of good performance with logistic regression, CART and perhaps GBM. This highlights that even thought the actual model used to fit the chosen features is the same in each case, the model used within RFE can make an important difference to which features are selected and in turn the performance on the prediction problem.
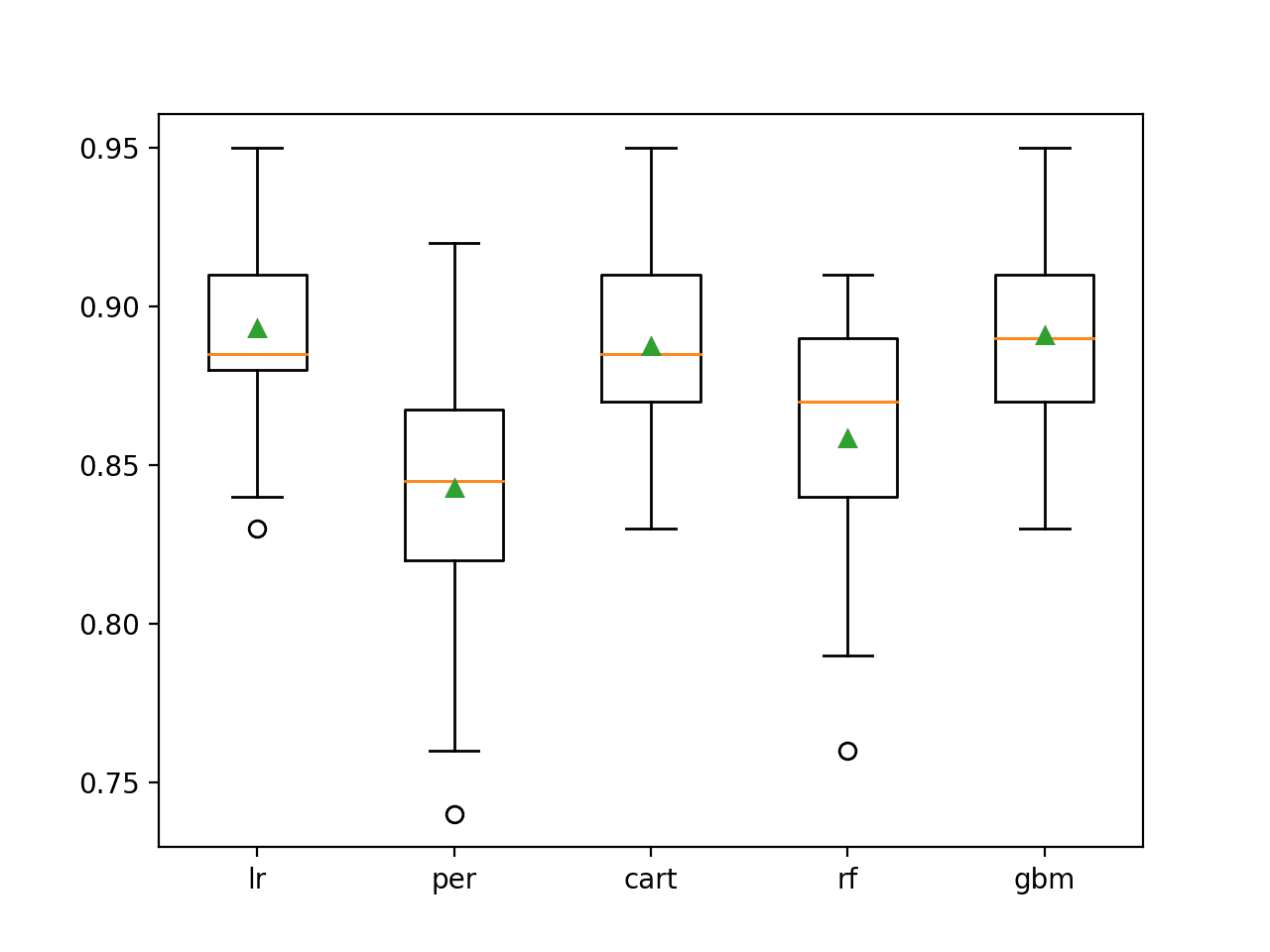
Box Plot of RFE Wrapped Algorithm vs. Classification Accuracy
Further Reading
This section provides more resources on the topic if you are looking to go deeper.
Tutorials
Books
- Applied Predictive Modeling, 2013.
Papers
APIs
- Recursive feature elimination, scikit-learn Documentation.
- sklearn.feature_selection.RFE API.
- sklearn.feature_selection.RFECV API.
Articles
Summary
In this tutorial, you discovered how to use Recursive Feature Elimination (RFE) for feature selection in Python.
Specifically, you learned:
- RFE is an efficient approach for eliminating features from a training dataset for feature selection.
- How to use RFE for feature selection for classification and regression predictive modeling problems.
- How to explore the number of selected features and wrapped algorithm used by the RFE procedure.
Do you have any questions?
Ask your questions in the comments below and I will do my best to answer.





Hi Jason, great tutorial on feature selection. Can this be applied to ordinal data as well?
My dataset contains wellbeing measures(mental health, nutritional quality, sleep quality etc.) as input features and the values for those can range from 0-10. I want to know the most useful features among them. Can I use this technique or some kind of correlation analysis?
Thanks!
Yes, RFE is agnostic to input data type.
Very nice tutorial on feature selection.
Thanks!
Hi Jason, thank very much for the tutor of RFE, finally I understand this topic. My question is: how can I see the variables that RFE is choosing in each fold of the cross validation? (in this situation I’m using StratifiedCV without repeat just 10 fold CV) i’m using your code the only different is that for select the variable I’m using Ridge and my model is RidgeClassifier. In the lines below you can see my code
rfe = RFE(estimator=Ridge(alpha=1.0000000000000006e-10), n_features_to_select=10)
model=RidgeClassifier(alpha=1.0000000000000006e-10)
Step=[(‘s’,rfe),(‘m’,model)]
pl=Pipeline(steps=Step)
cv=StratifiedKFold(n_splits=10, shuffle=True, random_state=1)
n_scores = round(cross_val_score(pl, X_uo, y_uo, scoring=’accuracy’, cv=cv,n_jobs=-1).mean(),3)
print(‘Stratified Cross validation Accuracy form Ridge model with all the variables: ‘, n_scores)
You cannot tell if you’re using cross_val_score() but you may want to look at cross_validate() which allows you to read more details.
Hello Jason. Can you please elaborate on the following? “When using cross-validation, it is good practice to perform data transforms like RFE as part of a Pipeline to avoid data leakage.”
Why using data transforms will avoid data leakage?
Good question.
If data from the test set is used or available to the model during training it is called data leakage:
https://machinelearningmastery.com/data-leakage-machine-learning/
This can give misleading results, often optimistic.
good explained
Thank you for your feedback and support abhi! We greatly appreciate it!
Thank you for the article. Where would you put cross-validation/model tuning?
1. Do cross-validation at the beginning using all the features and then perform RFE;
2. Use RFE and then do cross-validation using the model with the features selected;
3. Include cross-validation inside RFE: at each iteration in RFE tune a model using the current subset of features, remove the least important, perform cross-validation again using the new subset and discard the least important, and so on.
Which one is the correct approach?
Ideally CV is used to evaluate the entire modeling pipeline.
Within the pipeline you might want to use a nested rfecv to automatically configure rfe. We call this nested cross-validation.
how is this idea different from backward selection? it’s well known that either backward, forward and stepwise selection are not preferred when collinearity exist which is more prevalent these days with decent amount of variables.
It is very similar.
Brilliant, quite comprehensive. Thanks a lot, mate for your efforts.
Thanks!
Hi Jason, Thanks a lot for the nice explanations.
I would like to know, how to get the features selected after all models were tested. I mean, after a chose the winner model how I could get the winner features?
After the model is fit on your data, you can access the RFE object and report the selected features.
See the section “Which Features Were Selected”
Hi Jason,
Great tutorial.
Would you help me to understand why those selected column (2,3,4,6,8) in “Which Features Were Selected” are different from the previous RFE explore number of features where significant columns are (4-7)?
Thanks,
It is possible that features that RFE think are not important do in fact contribute to model skill.
Is it possible to extract final regression formula or equation from any successful prediction models like conventional regression models ? Thank Very Much.
Perhaps from a simple linear regression model, e.g. you can retrieve the coefficients from the fit model:
https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.LinearRegression.html
Is there any robust tutorial about nonlinear curve estimation with many input variables
Sorry, I don’t have a tutorial on this topic, hopefully in the future.
Hi Jason,
Nice post. In Section ‘RFE with Scikit learn’ you explained that RFE can be used with fit and transform method using ‘rfe.fit(X,y)’ and ‘rfe.transform(X,y)’. isn’t it be only ‘rfe.transform(X)’ without class labels? Thanks!
No, it is both input and output so subsets of features can be evaluated.
Also see this:
https://scikit-learn.org/stable/modules/generated/sklearn.feature_selection.RFE.html#sklearn.feature_selection.RFE.fit
Dear Dr Jason,
I had a go of applying the above to the iris data using the Pipeline and not using Pipeline.
Using Pipeline
This is application of the iris data without pipeline
From the results there is little difference between using the Pipeline and not using the Pipeline.
What is the point of implementing a Pipeline when there is little difference between the mean and stddev of the n_scores?
Thank you,
Anthony of Sydney
Dear Dr Jason,
Apologies, I used the mean(std). I should have used the mean(n_score) and std(n_score)
The same question applies – what is the point of the Pipeline, where it produces little differences in the results of the scores?
Thank you,
Anthony of Sydney
We use a pipeline to avoid data leakage:
https://machinelearningmastery.com/data-preparation-without-data-leakage/
Dear Dr Jason,
Thank you for referring me to the article on “data preparation without data leakage”.
Generally it is about the way the data is prepared. In the above ‘experiment’, the relevant headings are “Cross-Validation Evaluation With Naive Data Preparation” and “Cross-Validation Evaluation With Correct Data Preparation”.
I understand the pipeline method is to put the operations in a list.
The two lines from the ‘naive’ and ‘correct’ data preparation methods respectively are
Yes, the mean and stddev of the scores results were slightly different.
Please excuse my concept of leakage in computing. I thought ‘leakage’ meant something to do with garbage collection in C or Java. Obviously it is not the case in the data preparation tutorial
Question please:
How can an assignment of testing and training data leak into each other when you make an assignment of a variable to another variable, you wouldn’t expect that when you assign X_test and y_test to a k-fold operation to mix with the X_train and y_train.
Or put it another way: although pipelines are not the same as threads, if you don’t funnel a set of procedures in a certain order, you won’t get accurate answers, in the same way that if you don’t have threads the execution of a particular block of code you won’t get accurate answers?
Thank you,
Anthony of Sydney
It has nothing to do with threads or programming.
Instead, it has to do with making use of data by the model that it should not have access to. E.g. access to “information” from the test set when training the model. Perhaps re-read the tutorial on data leakage.
Dear Dr Jason,
Thank you for the reply. I have re-read and still have a ‘mental’ block’
Split this into background and questions.
Background:
Piplines have to do “…with making use of data by the model that it should not have access to…”
From the blog.
“….knowledge of the hold out test set leaks into the dataset used to train the model…”
“….information about the holdout dataset, such as a test or validation dataset, is made available to the model in the training dataset….”
“….This could happen when test data is leaked into the training set,…”
If I split the data into train and test, how does the information leak from the train to test or test to train set after I split the data.
Then once I split the data into train test, how does test or train data leak back?
Furthermore when I do the model fitting,
I am fitting X_train and y_train.
I don’t see how X_test, y_test leaks into X_train or y_train and vice versa.
And when data preparation is performed for using the MinMaxScaler() you are transforming the data using MinMaxScaler amongst the scalers MinMaxScaler, RobustScaler, StandardScaler and Normalizer, you want better convergence of the model.
In other words how does info from the X_train interfere with X_test and vice versa. Same question for leakage into y_train and y_test and vice versa.
In addition:
From the naive model, I don’t get how cv = RepeatedKFold causes leaks when cv is already assigned similarly, when the data is used in scores=cross_val_score(model,X,y,……..cv=cv….) that there is leaking.
From the Pipeline model
I don’t understand how information from X, y is leaked given that there is no split of X and y.
Questions/Summary – of leakage:
There are two methods
* one uses data preparation for faster convergence of model, BUT I don’t understand how transformation reduces the leakage when the data is already assigned.
* The other uses a Pipeline. Again I don’t understand how putting a pipeline with a list containing a sequence of commands causes interference between X and y.
I am sure that there is something simple that how a mere assignment of variables STILL causes leaks unless I either prepare the data and/or use pipelines.
Thank you,
Anthony of Sydney
You can leak from test to train if you scale train using knowledge of test, e.g. you normalize and calculate min/max using the entire dataset.
A pipeline ensures that the transforms are only ever fit on the training set.
Dear Dr Jason,
This is what I understand about based on your answer and the blog.
Under the heading “Train-Test Evaluation With Correct Data Preparation” and subheading “Tying this together, the complete example is listed below”, the anti-leakage preparation by transformation was performed on SEPARATE X_train and X_test. .NOT ON THE WHOLE X features.
As I understand it, the standard deviation of the X_train may not necessarily be the same as the standard deviation of the X_test, NEITHER WHICH ARE THE SAME as the std deviation of the whole X.
Then in order to make a prediction, you transform the values you wish to predict.
Thank you again, it is appreciated.
Anthony of Sydney
Yes, if we fit the transform on the whole dataset we get leakage. If we fit the transform on the training set only, we don’t get leakage.
Dear Dr Jason,
Thank you for that, it is appreciated.
Anthony of Sydney
You’re welcome.
Dear Dr Jason,
When I submitted the question, I had errors on the web browser due to a slow response. I submitted the same question at the bottom of the page. Please ignore the above submission as the same submission is asked at the bottom.
Anthony of Sydney
Hi Jason.
Thanks for the insight on RFE.
This question has probably been aske but I’m looking for clarification around the process of RFE using CV.
From the “exploring the number of features” section:
Let’s say we have training data and we split it into 5 folds. We will leave the testing data for now.
Fold 1-4 is used and we run using RFE on a range of 3 to 5 features to be selected. I believe it runs the model on the best 5 features, records a score on fold 5, runs the model for 4 features, records the score on fold 5 and does the same for 3 variables also. We then move onto using Fold 2-5 where we evaluate on fold 1 etc.
Am I right in saying that the 5,4 and 3 features chose in this fold can be different from those chose in the other folds we are training on?
If that is the case, when we go to use the entire training data to create the model and run on the holdout/unseen training data, how does the model choose what features to use if it is returning different features on every fold across the 3, 4 and 5 features selected?
great!
Thanks!
Thanks Jason, Really ueful stuff!!
I have one questions. Once you run feature selection, cross validation and grid search through pipeline, how do you access the best model for predictions on x_test?
You fit a new final model using the techniques and configuration you discovered work best on your dataset:
https://machinelearningmastery.com/train-final-machine-learning-model/
Thanks Jason, Yes that makes sense to me. But I am not sure how do I access selected features when I use ‘cross_val_score’ and the ‘pipeline’ in a loop (as you show in “RFE for Classification”).
What I am trying to do through a loop is:
1. For every combination of hyperparameters and RFE, run a model fit and cross_val_score
2. Select best features with reference to this model and transform inputs
3. Fit a new model using selected features only and use it to predict with test data
4. Check accuracy, so that in a box plot I can also visualise for every model run how it performed on the test data.
I have completely independent validation data that I would use at the end for independent validation for the ‘best model’.
But I don’t know how I get selected features after calling cross_val_score.
Thanks!!
You don’t need to access the features as RFE becomes part of your modeling pipeline.
You can select the features chosen by RFE manually, but the point is you don’t need to. RFE does it for you.
Nice article. Thanks
1. When we use RFECV to Automatically Select the Number of Features then how can we know what features are selected using RFECV method?
2. Can we first use RFECV and then do cross-validation using the model with the selected features from RFECV?
You can run the method manually if you like and have it print the features it selected.
Yes.
Thanks. You mean manually means using RFE method?
I mean that you can run the RFE or RFECV method in a standalone manner and review what it is doing.
Sure. Thanks a lot.
Very informative article. Thank you for posting.
I have a question related to having a mixed data set i.e it has both numerical and categorical inputs and requires a categorical output. If I split the data set into two files one containing the numerical data and another containing the categorical data, and then run the appropriate feature selection method (eg ANOVA and Chi Squared) on each data set, then is it appropriate to use the information obtained on the most important features of each data type, to alter the original data set to select the appropriate fields? I am wondering if this is appropriate or if it introduces bias?
Thanks!
Try it and see if it performs better than an RFE or using all features.
It is bias, hopefully can find a way to do it within a cv fold.
Dear Dr Jason,
The code under subheading “Explore Base Algorithm”, the lines in particular are:
lines 25-27. This is similar to the code used in your book, listing 15.21 p187
We have the RFE select up to five features using LogisticRegression.
How does the DecisionTreeClassifier work with the RFE(LogisticRegression).
Thank you,
Anthony of Sydney
The decision tree will take the features selected by the RFE and fit a model.
The logistic regression model is only used by the RFE to evaluate different subsets of features selected by the RFE.
Dear Dr Jason,
Thank you for the elaboration. It is appreciated.
Anthony of Sydney
You’re welcome.
Dear Dr Jason,
Again thank you for the reply. I would like to apply your reply to listing 15.21 on page 186 (203 of 398) of “Data Preparation For Machine Learning” a book I highly recommend.
Applying your previous answer in order to get a better understanding:
The first line, rfe selects features using differerent subsets based on the DecisionTreeClassifier. The selected features are evaluated in the DecisionTreeClassifier = model.
Thank you,
Anthony of Sydney
Yes, it can be a good idea to use the same model within RFE as in following RFE.
Dear Dr Jason,
Thank you, it has answered my question, it is appreciated.
Anthony of Sydney
Thanks for sharing this. Your blog is better that sklearn documentation 🙂
Quick question: When I tune n_features_to_select parameter and use a DecisionTreeClassifier as estimator I get similar results to yours, however when I use a LogisticRegression instead, I always get the same results, no matter the value of n_features_to_select is.
With DecisionTreeClassifier: (f-score)
>2 0.742 (0.009)
>3 0.742 (0.009)
>4 0.741 (0.009)
>5 0.741 (0.009)
>6 0.741 (0.009)
>7 0.740 (0.010)
>8 0.739 (0.010)
>9 0.739 (0.010)
With LogisticRegression: (f-score)
>2 0.742 (0.009)
>3 0.742 (0.009)
>4 0.742 (0.009)
>5 0.742 (0.009)
>6 0.742 (0.009)
>7 0.742 (0.009)
>8 0.742 (0.009)
>9 0.742 (0.009)
What could be the reason ?
Maybe it is not working since it is part of a Pipeline?
Thanks
Thanks!
Perhaps there is a bug in your logistic regression example?
Perhaps varying the features does not impact model skill (unlikely)?
Hi Jason,
Your articles are a great source of information. Have one question, if I understand it correctly, for using RFE, we need to at first normalize or standardize the available data in order to get the correct features according to the importance in the model. Else, the features with smaller values will have a higher coefficient associated and vice versa. This is my understanding. Please correct me if I am understanding it wrong.
Thanks
Thank you!
It depends on the model you are using. If you are using a tree within RFE, then no. If you are using a logistic regression, then probably yes.
Hi Jason,
Your articles are a great source of information
Thanks!
Hi Jason,
When tuning the best of number of features to be selected by rfe, shouldn’t we drop duplicates before running the model ?
i.e. if you keep only 2 variables, you will probably have more duplicated rows than if you use 5.
This can happen with categorical features or a mixture between categorical / numerical
Thanks a lot 🙂
Data cleaning like removing duplicates rows and columns can help in general, see this tutorial:
https://machinelearningmastery.com/basic-data-cleaning-for-machine-learning/
can we plot RMSE(root mean squared error) using the RFE algorithm?
as may not be as it is float
how can i plot then ?
How would you plot it exactly?
what algorithms that can be used in the core RFE for regression and how to calculate accuracy for these algorithms thank you.
Any regression algorithms you like.
when try to use algorithms that can be used in the core RF for regression problem not classification i get this error
ValueError: Unknown label type: ‘continuous’
what can I change in the code to avoid this error
Sorry to hear that you’re having problems, perhaps start with the regression example above and adapt it for your project?
Hi Jason is there a way to see which features were selected during the cross validation instead of fitting in all the data to get these?
It seems to me that the accuracy you obtained in the section “Automatic Select the Number of Features” was not based on the features you obtained in the section “”Which features were selected?”. The first section uses CV and the second fits in all the data.
Yes, for each fold if you enumerate manually and print the features selected by the object.
But why do you need to know? Do you care what coefficients a linear regression model chooses each fold? Never!
I mean how can we extract the subset selected that outputs that cross-validation score.
sorry , that was a question.
You can do it by manually enumeration the cv folds and evaluate your model, then access the RFE within the pipeline and print the reported features for that fold.
But I am asking why do you want this information? It is irrelevant!
I thinking of it for interpretation. To know which 10 features were found as the most important ones.
You can run the model once in a standalone manner to discover what features might be important.
But please let me know if there is a better way. Thanks for your help
Yes, you can run the procedure on a train/test split of the data to learn more about the dataset.
This is a separate procedure from evaluating modeling pipelines where knowing what features were selected does not change anything.
Hi Jason,
Thank you for the useful blog. Can you please tell me that, for a regression problem, if I can use the “DecisionTreeRegressor” as the estimator inside the RFE and “Deep Neural Network” as the model? If not, do you have any recommendation for the estimator inside the RFE when I want to select the best subset of data for a DNN algorithm?
Thanks in advance,
Masoud
Sure.
Generally, it is a good idea to use the same algorithm in both cases, but it is not a rule.
Thank you dear Jason.
You’re welcome.
Thank you, Jason, for the very informative blog post.
I have a few questions to discuss.
In the conventional method that the statistician uses to fit the regression model.
They would do a single feature test for p-value and select only those variable which tends to have significant, such as p<0.15, for first-round filtering features.
Then, they would use multiple regression and using forward or backward elimination for the final feature selection.
Do you have information on which one gave a better result? Comparing single RFE alone, or perform the single feature filter first before RFE, or RFE vs forward/backward elimination.
There is no best method:
https://machinelearningmastery.com/faq/single-faq/what-feature-selection-method-should-i-use
Is it worthwhile doing RFE when using more complex models, such as XGBoost?
Yes, if you are not using xgboost.
Hello Jason,
first of all thanks for all your amazing work! I gained so much knowledge through your website. I can’t put into words how much I thank you for that.
I implemented your pipeline on an own dataset. When I want to check on the different feature importances all 47 features are equally important. Looks like I got some leakage, doesn’t it?
You’re welcome.
Maybe, or maybe the technique cannot tell the difference between your features – e.g. your problem might not be predictable.
Hi Jason
I have a question. Say we are using RFE but we haven’t chosen a specific model to predict our output. As in we have a list of possibilities, whether that is SVM, Gradient Boost, or Random Forest etc (classification but also a different list for regression). When we perform cross-validation on RFE and set it up to automatically pick the number of features, would we have to repeat it for every model?
Also you used DecisionTree as both the estimator and the model, can we use different models but keep the DecisionTree as the estimator? Ex) Estimator=DecisionTree Model=SVM
Can we also use different estimators?
Ahhh. I missed the end. Thanks for the article. I should finish reading before asking questions.
Follow up:
When doing feature selection and finding the best features from using RFE with cross-validation, when we test other ML algorithms for the actual modeling of the data, would we run into the issue that different models will work better with different chosen features?
At the end of the article you compared different estimators with the same model. Say we pick the best one but later we still have to optimize the hyperparameters of the model using Gridsearch. How do we know that this is still the best model for us? How do we know that the other estimator/model combinations couldn’t be better if we optimized with grid search the hyperparameters in the model? At what point are we able to stop with that peace of mind?
Yes.
Test everything, use what results in lowest error.
Stop when the results are good enough, or when you run out of ideas, or when you run out of time.
No problem.
You can use different models inside and outside if you like, using the same model might be preferred.
Each, when considering a suite of models, each should be considered in the same modeling pipeline (applying transforms to the data like rfe)
Would we remove highly correlated features before applying RFE?
Probably.
It depends on the model you’re ultimately using. Also it depends if you have tons of features and are looking for ways to speed up your pipeline.
# define dataset
X, y = make_classification(n_samples=1000, n_features=10, n_informative=5, n_redundant=5, random_state=1)
# create pipeline
rfe = RFECV(estimator=DecisionTreeClassifier())
model = DecisionTreeClassifier()
pipeline = Pipeline(steps=[(‘s’,rfe),(‘m’,model)])
# evaluate model
cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1)
n_scores = cross_val_score(pipeline, X, y, scoring=’accuracy’, cv=cv, n_jobs=-1, error_score=’raise’)
# report performance
print(‘Accuracy: %.3f (%.3f)’ % (mean(n_scores), std(n_scores)))
I want to know how many features by RFECV but since it is in pipeline object I am not able to get the support and rank attribute. Please help!!
If you run RFE on a train set only you can see the features that were selected – as we do in the above tutorial.
When used as part of a pipeline, we don’t care what was selected, just like we don’t care what the specific decision tree looks like – only that it performs well.
Hey,
Thanks for such a nice tutorial.
I am not able to understand one thing. We use RFE inside pipeline and then use gridsearchCV to find out optimal number of features let’s say [2,5,10].
So my understanding is, gridsearchCV will split the data into k folds. Use k-1 subsets for training and apply RFE on it to select best performing features. Finally evaluate it on remaining subsets to evaluate it. Repeat the same step k times to find out the average model performance. Follow the same procedure for each value for RFE features.
My question is, does RFE select same features in each fold or they could be different. If the later is true, why gridsearchCV returns only one list of selected features for best performing parameters/model.
Thank you in advance.
RFECV will select the number of features for you, no need to grid search as well.
Thank you, Jason, for the very informative blog post.
I have a question.When I use RFECV, why I get different result for each run.Sometime return 1 feature to select, sometime return 15 features.Thank you so much.
You’re welcome.
Good question, see this:
https://machinelearningmastery.com/faq/single-faq/why-do-i-get-different-results-each-time-i-run-the-code
Hi Jason,
Thank you for the articles. I have read few of them about feature selection.
I have a dataset with many numerical and categorical variables.
I am trying to check which features have a significance w.r.t. to the target variable (binary).
I tried various approaches, separately, using numerical features used ANOVA and for categorical I used chi-squared. In addition, I wanted to use RFE to compare my results.
When I use the below:
rfe = RFE(estimator=DecisionTreeClassifier(), n_features_to_select=7)
rfe.fit(X4,y4)
It returns an error – could not convert string to float: ‘ – ‘.
hence, my question is, do I need to encode the categorical features? Also, when I check the datatype of the categorical variables, it is seen as float.
Please let me know. Thank you in advance.
You’re welcome.
Yes. You must ordinal encode your categorical variables prior to using RFE.
Thanks Jason. I will encode my categorical variables. I thought RFE would take care of them automatically for me 🙂
Another question, if my goal is to know which features have a significance w.r.t to the target variable rather than prediction, I think by using RFE or any of the other filter approaches such as chi-squared, ANOVA should be sufficient. I don’t think I need to create a model, however please let me know if my understanding is incorrect.
Also, are there other approaches to find which features are significant w.r.t to the target variable?
No problem.
You could calculate a statistical test for each pair of input and target variables and compare their results. Significance may or may not correlated with best performing.
Yes, this may help:
https://machinelearningmastery.com/feature-selection-with-real-and-categorical-data/
Thank you Jason.
Sorry, I have one more question. I am stuck with my issue.
I was trying to encode my categorical variables in my dataset, however I am not sure how to get them back in the same dataframe. For example- in my dataframe, 2 features are categorical, 3 are nominal, when I use OrdinalEncoder or Onehotencoder, it expects categorical columns only. Hence, when it finishes encoding, how do I retain the encoded columns and 3 nominal features? I read your article about columntransformer as well, however as I am not creating a model, only want to use RFE, how do I proceed? Your example uses a pipeline approach, hence not sure how to proceed.
Good question, you can use a columntransformer:
https://machinelearningmastery.com/columntransformer-for-numerical-and-categorical-data/
Hi Jason, thank you for the awesome post! I have a question when using sklearn MLPRegressor as a model in RFE. It looks like RFE cannot handle neural network as a model, is that true? as I am getting this error message when running MLPRegressor model in RFE.
###RuntimeError: The classifier does not expose “coef_” or “feature_importances_” attributes###
Please help…. Thank you in advance !
I don’t know sorry. I have not tried.
Hi Jason, could you also please advice me on what feature selection method I should use if I have a regression problem with multiple outputs. As almost all of the future selection methods you wrote, filter-based and wrapper are only for multiple inputs vs one output.
Thank you 🙂
Perhaps try RFE, perhaps try some of the regression methods described here:
https://machinelearningmastery.com/feature-selection-with-real-and-categorical-data/
Hi Jason, this was super helpful, thank you! I had a question. If using in classification, how do we know the performance metric that the wrapped classifier uses to judge performance and therefore feature importance? The reason I ask is, if we were to have imbalanced classes if the wrapped classifier uses ‘accuracy’, the most important features may not actually be the ones we are after, rather we would want to use, for example AUC. Can you shed any light on this? Thank you! Tash
It’s simpler than that.
The least important features are removed based on the importance of those features as determined by the inner model. Not the performance of the inner model on the dataset.
Hi Jason,
When evaluating the MAE, shall we take the STD into consideration? Therefore, we may discard the configuration with the smallest MAE in case the correspondent std is very high compared to the rest.
Thanks in advance.
If you are computing the mean MAE then reporting mean MAE and std MAE is a good idea. Otherwise no.
Hi Jason , can you explain how logistic regression gives importance score to each feature.
Thanks
The coefficients can be directly interpreted as variable importance scores. Or at least the abs() values can be.
Okay, Thanks.
So what if we are working in unsupervised setting in which there are no labels, then which estimators can be used?
Also, can we use autoencoder in this case for feature selection?
Sorry, I don’t know much about feature selection for unsupervised learning – I guess it depends on the type of unsupervised learning problem.
If there is no target variable, then feature selection via statistical correlation or by subset evaluation cannot be used.
An autoencoder will perform a type of automatic feature extraction, perhaps that is useful for you.
How to apply RFE for multiple output like
X = data.drop([‘Mi’,’P’, ‘T’], axis=1)
y = data[[‘Mi’,’P’, ‘T’]]
and how to evaluate this kind of model
I’m not sure RFE supports multiple outputs.
Perhaps try it and see?
Hi Jason,
Thank you so much for this great article.
I had two questions; Can we use RFE method for a dataset that contains only categorical variables (70 variable)? Also, some of these variables are ordinal. Should we one hot encode these ordinal variables before or after RFE?
Thank you in advance.
Yes.
Perhaps try an ordinal encoding as a first step.
Hey Jason,
Great article. I have been following your work for quite a while. I have been looking for some techniques to find out relevant/important features from a dataset containing more than 18,000 variables(continuous). Since RFE will take years to compute that. Can you please suggest any technique which I can try?
Thank you in advance.
Thanks!
Perhaps start with a fast/simple linear method like correlation with the target.
Hi Jason,
Suppose,
If I choose 10 features by taking 2, 3, 4… 9 features separately using for loop i (2, 9) and finally print their results for k-fold cross validation to see their performance.
How can I get the selected features name for each model? Suppose, for taking 3 features, which three feature is selecting here among 10? How can I get those feature name?
Please let me know.
Regards
Ashiq
You can re-run RFE with a train-test split and then use the array indexes reported by RFE into an array of column name or map them manually.
See the section “Which Features Were Selected” above.
Thanks for the tutorial.
Excuse me, I am new to Python, but how to get the features selected from the pipeline?
See the section “Which Features Were Selected”.
Hi jaon! I have a question. How can we apply RFE with HistGradientBoostingRegressor? It gives an error since probably we cannot get features importances directly using HistGradientBoostingRegressor!
REFE can be used with HistGradientBoostingRegresso directly as far as I know, perhaps you have a bug in your code.
Perhaps these tips will help:
https://machinelearningmastery.com/faq/single-faq/can-you-read-review-or-debug-my-code
hello sir please answer REF is used as feature selection and then decision tree model suits for thyroid disease daignose or any one good model recommended? thanks in advance
Perhaps try it and see if it is effective on your dataset and compare to other methods.
Everyone looking for SHAP for feature selection (and hyperparameters tuning), I suggest shap-hypetune (https://github.com/cerlymarco/shap-hypetune).
A python package for simultaneous Hyperparameters Tuning and Features Selection for Gradient Boosting Models.
It supports feature selection with RFE or Boruta and parameter tuning with Grid or Random Search.
Thanks for sharing.
Hi Jason,
nice tutorial about RFE as a wrapper-type feature selection algorithm!.
One question, related to your section exploring number of features. When RFE perform the features selection through the argument ‘n_features_to_select’ it is always retaining the best ones?.
I mean, I can not obliged to RFE to use a specific ‘subset’ of features when defining the total amount of ‘n_features_to_select?.
Anyway my experience is I do not feel so much confortable using RFE ‘facility’:
– you can not use it for all of the keras model implementation, besides others Sklearn model that does not support ; coef_, feature_importances_ as fit information.
– the features selected results are some time weirds for certain estimators (models)
I rather prefer to use “permutation_importance, as a more robust function (even from Sklearn) and more flexible to use with more models (e.g. keras model wrapper) and I think is a better performer
thanks
Correct, it will be forced to use the best n features specified.
sir please guide how can i compare ? please respond
Compare what exactly?
What a great article! Fascinated by sklearn.feature_selection, I tried it out on kaggle’s titanic_survivors, but the three principal features I asked to be selected were not the same going forward or backward. I would be interested to understand why this happens.
A change to the algorithm will result in changes to the features selected as comparisons between features are relative and stochastic/noisy.
Hi Jason, great tutorial.
Could you please help me with something? Ok, you find what K (for example K=3) is best using CV. Now, what we need to do is retrain using all data (except for the test set) using that K (K=3). Then, after evaluating the performance in test, we retrain again using training + test sets, again using K=3, and that one is your final model (the one you put in production).
Here is my question. Why everybody is so interested in knowing what were the selected features? The way I see it is that actually we don’t care about what variables were selected (when we use CV we care about the pipeline, not about the specific of what variables are selected in each fold; we want the process to be robust, that’s all). Even within a particular iteration in the CV with K=3, different variables are selected each time, since each iteration use a different dataset!
Actually, it could happen that if K=3 was the best (and the feature selected were A, B and C when training with all data (training + test)), that changes when we get new data and retrain again (we could keep K=3, but now, since we have more data, and thus a different dataset, now the selected features might be B, E and F). And in fact that would perfectly fine, since we only care about the pipeline, not the specific… right???
Great article! If I was comparing different models would I want to use the same estimator for each model within the RFECV to have the same predictors each time or explore the ‘best estimator for within RFECV for each model?
You need the same metric for an apple-to-apple comparison. One example, you cannot compare RMSE to MSE numerically because one is the square root of another. RFE tells for the same output, what subset of input can help the model do the best. Bear this in mind, you should understand what to do for the CV so you can make the comparison.
Keep in mind that SVM-RFE is a patented technology. Usage of SVM-RFE without license (writing your own or using the one written into SKLEARN) will open you up to infringement liability. Ask Intel. They are being sued by Health Discovery.
Thanks for sharing. I am no lawyer here but I can find the patent document and it seems to be expired last year: https://patents.google.com/patent/US8095483B2/en
Is it possible to apply on KNN, SVM, Bagging etc..
Why not?
Thanks, Jason, for your awesome work, as always!
I have a quick question, please.
How do we know how many “best” features to select?
You don’t know. That’s more like a trade off that you need experiments to confirm.
Hi Jason,
Can we use ANN model as estimator in RFE. When i tried giving ANN as estimator, it gave me following error: AttributeError: ‘Sequential’ object has no attribute ‘_get_tags’
Any idea regarding this?
This post will help: https://machinelearningmastery.com/use-keras-deep-learning-models-scikit-learn-python/
Hi Jason,
Here, you used Decision Tree. Is it possible to apply other types like KNN, XGBooster in RFE?
Thanks. While trying to print this for study, it looks some contents are overlapped, not clear. Please advice how to print.
Hi Meenakshi…The blog content is not optimized for printing purposes.
Dear Jason,
Thank you for this helpful post.
I used this code for a regression problem. I am getting negative values for mean like below:
>2 -8.26728 (0.21538)
>3 -5.32587 (0.29661)
>4 -3.35181 (0.41920)
>5 -2.82741 (0.39231)
>6 -2.29025 (0.35978)
>7 -2.09409 (0.27335)
>8 -1.93794 (0.19657)
>9 -1.97243 (0.22962)
>10 -1.93483 (0.18773)
Are my values right or I should only get positive mean values? and if it is right what number of features are the best one? the one corresponding the 8 features?
Hi Kelly…Some model evaluation metrics such as mean squared error (MSE) are negative when calculated in scikit-learn.
This is confusing, because error scores like MSE cannot actually be negative, with the smallest value being zero or no error.
The scikit-learn library has a unified model scoring system where it assumes that all model scores are maximized. In order this system to work with scores that are minimized, like MSE and other measures of error, the sores that are minimized are inverted by making them negative.
This can also be seen in the specification of the metric, e.g. ‘neg‘ is used in the name of the metric ‘neg_mean_squared_error‘.
When interpreting the negative error scores, you can ignore the sign and use them directly.
You can learn more here:
Model evaluation: quantifying the quality of predictions
Is this feature selection using RFE possible only for the model with as feature_importances_ or coef_ internal property?
If so, I think we can’t use this method for MLPRegressor (MLPR) or HistGradientBoostingRegressor (HGBR) because they don’t have those properties.
Do you know how we can add new functions like feature_importances_ to MLPR, HGBR?
Hi ykcho…The following resource may add clarity:
https://towardsdatascience.com/powerful-feature-selection-with-recursive-feature-elimination-rfe-of-sklearn-23efb2cdb54e
Hi James. I just want to double check. You said the article may add clarity.
But, in that article, only RandomForest model is considered and it is said all sklearn models have ‘coef_’ or ‘feature_importances_’. But, as far as I know, it is wrong. The models which I mentioned (MLPR, HGBR) do not have those attributes.
So, I want to ask you your meaning for clarity, again.
Hi James. I just want to double check. You said the article may add clarity.
But, in that article, only RandomForest model is considered and it is said all sklearn models have ‘coef_’ or ‘feature_importances_’. But, as far as I know, it is wrong. The models which I mentioned (MLPR, HGBR) do not have those attributes.
So, I want to ask you your meaning for clarity, again.
RFE is not a good wrapper method.
RFE is a bit of a hybrid. It looks and acts as a wrapper, similar to backward selection. But its main drawback is its selection of variables is essentially univariate. It uses a univariate measure of goodness to rank order the variables. In this sense it behaves like a filter.
I like this description of feature selection methods:
Filters: fast univariate measure process that relates a single independent variable to the dependent variable. A filter can be linear (e.g., Pearson correlation) or nonlinear (e.g, univariate tree model). Filters typically scale linearly with the number of variables. We want them to be very fast , so we use a filter first to get rid of many candidate variables, then use a wrapper.
Wrappers: called such because there is a model “wrapped” around the feature selection method. A good wrapper is multivariate and will rank order variables in order of multivariate power, thus removing correlations. Wrappers build many, many models and scale nonlinearly with the number of variables. It’s best to use a fast, simple nonlinear model for a wrapper (eg, single decision tree with depth about 5, random forest with about 5 simple trees, LGBM with about 20 simple trees). It really doesn’t matter what model you use for the wrapper (DT, RF, LGBM, Catboost, SVM…). as long as it’s a very simple nonlinear model. After you do the proper wrapper the result is a sorted list of variables in order of multivariate importance. You don’t get this with RFE.
In practice you might create thousands of candidate variables. You then do feature selection to get a short list that takes correlations into account. You first do a filter, a univariate measure, to get down to a short list of maybe 50 to 100 candidate variables. Then you run a (proper) wrapper to get you list sorted by multivariate importance, and you typically find maybe 10 to 20 or so variables are sufficient for a good model. Then you use this small number for your model exploration, tuning and selection.
Sure, many nonlinear models by themselves will give you a sorted list of variables by importance, but it’s impractical to run a full complex nonlinear model with hundreds or thousands of candidate variables. That’s why we do feature selection as a step to reduce the variables before we explore the final nonlinear models.
RFE/RFECV is a poor stepchild in between these. Its variable ranking is essentially univariate, like a filter. It doesn’t remove correlations, also like a filter. But it has a model wrapper around it, so it looks like a wrapper. It decides how many variables are ranked #1, sorts the rest by univariate importance, and doesn’t sort the #1 variables (or any variables) by multivariate importance.
My opinion: RFE is a popular but lousy wrapper. Use sequentialfeatureselector.
Thank you for your feedback steve c! Let us know if we can clarify any items from our blogs and/or other content.
Hi Jason,
many thanks for this article as well as all of your great content!
I have also read your “Feature Importance and Feature Selection With XGBoost” article, and have used both methods (RFE and the feature_importances_ XGBoost function) to the identify the most influential features for a given data set for which I am creating a regression model. For the RFE method, I take XGBoost as the estimator. The two methods end up identifying different features. E.g. I ask each for the top 10 features and only about half of them are the same ones. The others are different. Even though I use two methods that calculate differently, I would expect more similarity in the results, since both methods have the same purpose. (and especially since I use also XGBoost as the estimator for the RFE method)
Do you have an explanation for this?
Thanks!
Hi Sam…Thank you for your feedback! Did you utilize code directly from the tutorial or did you write your own?
Hi Jason, thank you very much for your article.
I have a question, should RFE and RFECV give the same or similar results? In my case, RFECV performed worse than RFE.
Hi Tati…You are very welcome!
RFE(Recursive Feature Elimination) andRFECV(Recursive Feature Elimination with Cross-Validation) are feature selection techniques that are available in thesklearnlibrary. They both aim to recursively remove features and rank them based on the performance of a given estimator. However, there are some key differences:1. **Cross-Validation**:
–
RFEsimply eliminates features based on the importance derived from a model. It does not use cross-validation by default.–
RFECVperforms feature elimination with cross-validation. At each step, it removes a feature and calculates the cross-validated performance to determine if the removal of the feature was beneficial.2. **Stability**:
– Because
RFECVuses cross-validation, it tends to provide a more stable and robust set of selected features compared toRFE, especially when the dataset has some variance or noise.3. **Computation Time**:
–
RFECVcan be computationally more expensive thanRFEbecause it uses cross-validation, which means fitting the model multiple times for each subset of features.4. **Result**:
– Due to the cross-validation step in
RFECV, the features it selects might be different fromRFEif the dataset has noise or if the model’s performance varies with different train-test splits.In summary, while both methods aim to eliminate less important features,
RFECVprovides a more robust feature ranking by using cross-validation. However, this comes at the cost of increased computation time. Depending on the specific dataset and problem at hand,RFEandRFECVmay give similar or different results. It’s always a good idea to validate the selected features on a separate hold-out set or using some external validation criteria.Hello, thank you for this post.
I have tried RFE and the result show that the feature subset with the highest accuracy was 21. Later, I attempted using all features for classification. Surprisingly, the feature subset of 21 did not turn out to be the one with the highest score during the classification process. Do you know why this happened?
Hi hartati…You are very welcome! There may be differences from each trial. What are your average findings over several trials?
How can we select the most important features based on recursive feature elimination without actually mention the amount of features to select?
Hi Yustina…The following resources may be of interest:
https://machinelearninginterview.com/topics/machine-learning/recursive-feature-elimination-for-feature-selection/
https://scikit-learn.org/stable/modules/feature_selection.html
Also, depending upon your application, deep learning methods may provide excellent performance without extensive feature elimination.
Hi,
I am trying to apply RFE in a problem of my own. Briefly, I have 92 observations each of which belong to one of 6 classes. For each observation, I have 610 features (in this case concentrations of various chemicals).I would like to select the features (chemicals) that are most uniquely present in each of the six classes. I first converted this dataset into a binary classification problem for each of the 6 classes (1= class of interest; 0=all other classes). I then observed that there were several features that were unique to the class of interest and absent from all other classes – I would assume these features would be important to the model, however, I also found that these features were highly correlated to each other. Nevertheless, I trained random forest, SVC, and logistic regression models on the dataset, and then ran RFE (as it has been suggested that RFE would be able to deal with correlated variables; specifically I ran it with RFE from sklearn). However, when I examine the features selected by RFE, it does not totally make sense. First, RFE seems to select features that are uniquely present in the class of interest (this makes sense). It also selects features that are uniquely absent in the class of interest (also kind of makes sense). But it also selects features that are present sporadically in the other (0) class and not present in a single observation in the class of interest. This makes no sense to me as there are other features that are uniquely present/absent in my class of interest. More importantly, these “weird” features are selected across two or more of my selected classifiers. Is there something wrong with my method? Are there other things I should try? Should I try to clean my dataset of correlated features prior to running RFE, even though I thought RFE can deal with correlated features?
Hi Tara…Please narrow your query to a single question so that we may better assist you.