It can be challenging to develop a neural network predictive model for a new dataset.
One approach is to first inspect the dataset and develop ideas for what models might work, then explore the learning dynamics of simple models on the dataset, then finally develop and tune a model for the dataset with a robust test harness.
This process can be used to develop effective neural network models for classification and regression predictive modeling problems.
In this tutorial, you will discover how to develop a Multilayer Perceptron neural network model for the ionosphere binary classification dataset.
After completing this tutorial, you will know:
- How to load and summarize the ionosphere dataset and use the results to suggest data preparations and model configurations to use.
- How to explore the learning dynamics of simple MLP models on the dataset.
- How to develop robust estimates of model performance, tune model performance, and make predictions on new data.
Let’s get started.

How to Develop a Neural Net for Predicting Disturbances in the Ionosphere
Photo by Sergey Pesterev, some rights reserved.
Tutorial Overview
This tutorial is divided into four parts; they are:
- Ionosphere Binary Classification Dataset
- Neural Network Learning Dynamics
- Evaluating and Tuning MLP Models
- Final Model and Make Predictions
Ionosphere Binary Classification Dataset
The first step is to define and explore the dataset.
We will be working with the “Ionosphere” standard binary classification dataset.
This dataset involves predicting whether a structure is in the atmosphere or not given radar returns.
You can learn more about the dataset here:
You can see the first few rows of the dataset below.
|
1 2 3 4 5 6 |
1,0,0.99539,-0.05889,0.85243,0.02306,0.83398,-0.37708,1,0.03760,0.85243,-0.17755,0.59755,-0.44945,0.60536,-0.38223,0.84356,-0.38542,0.58212,-0.32192,0.56971,-0.29674,0.36946,-0.47357,0.56811,-0.51171,0.41078,-0.46168,0.21266,-0.34090,0.42267,-0.54487,0.18641,-0.45300,g 1,0,1,-0.18829,0.93035,-0.36156,-0.10868,-0.93597,1,-0.04549,0.50874,-0.67743,0.34432,-0.69707,-0.51685,-0.97515,0.05499,-0.62237,0.33109,-1,-0.13151,-0.45300,-0.18056,-0.35734,-0.20332,-0.26569,-0.20468,-0.18401,-0.19040,-0.11593,-0.16626,-0.06288,-0.13738,-0.02447,b 1,0,1,-0.03365,1,0.00485,1,-0.12062,0.88965,0.01198,0.73082,0.05346,0.85443,0.00827,0.54591,0.00299,0.83775,-0.13644,0.75535,-0.08540,0.70887,-0.27502,0.43385,-0.12062,0.57528,-0.40220,0.58984,-0.22145,0.43100,-0.17365,0.60436,-0.24180,0.56045,-0.38238,g 1,0,1,-0.45161,1,1,0.71216,-1,0,0,0,0,0,0,-1,0.14516,0.54094,-0.39330,-1,-0.54467,-0.69975,1,0,0,1,0.90695,0.51613,1,1,-0.20099,0.25682,1,-0.32382,1,b 1,0,1,-0.02401,0.94140,0.06531,0.92106,-0.23255,0.77152,-0.16399,0.52798,-0.20275,0.56409,-0.00712,0.34395,-0.27457,0.52940,-0.21780,0.45107,-0.17813,0.05982,-0.35575,0.02309,-0.52879,0.03286,-0.65158,0.13290,-0.53206,0.02431,-0.62197,-0.05707,-0.59573,-0.04608,-0.65697,g ... |
We can see that the values are all numeric and perhaps in the range [-1, 1]. This suggests some type of scaling would probably not be needed.
We can also see that the label is a string (“g” and “b“), suggesting that the values will need to be encoded to 0 and 1 prior to fitting a model.
We can load the dataset as a pandas DataFrame directly from the URL; for example:
|
1 2 3 4 5 6 7 8 |
# load the ionosphere dataset and summarize the shape from pandas import read_csv # define the location of the dataset url = 'https://raw.githubusercontent.com/jbrownlee/Datasets/master/ionosphere.csv' # load the dataset df = read_csv(url, header=None) # summarize shape print(df.shape) |
Running the example loads the dataset directly from the URL and reports the shape of the dataset.
In this case, we can see that the dataset has 35 variables (34 input and one output) and that the dataset has 351 rows of data.
This is not many rows of data for a neural network and suggests that a small network, perhaps with regularization, would be appropriate.
It also suggests that using k-fold cross-validation would be a good idea given that it will give a more reliable estimate of model performance than a train/test split and because a single model will fit in seconds instead of hours or days with the largest datasets.
|
1 |
(351, 35) |
Next, we can learn more about the dataset by looking at summary statistics and a plot of the data.
|
1 2 3 4 5 6 7 8 9 10 11 12 |
# show summary statistics and plots of the ionosphere dataset from pandas import read_csv from matplotlib import pyplot # define the location of the dataset url = 'https://raw.githubusercontent.com/jbrownlee/Datasets/master/ionosphere.csv' # load the dataset df = read_csv(url, header=None) # show summary statistics print(df.describe()) # plot histograms df.hist() pyplot.show() |
Running the example first loads the data before and then prints summary statistics for each variable.
We can see that the mean values for each variable are in the tens, with values ranging from -1 to 1. This confirms that scaling the data is probably not required.
|
1 2 3 4 5 6 7 8 9 |
0 1 2 ... 31 32 33 count 351.000000 351.0 351.000000 ... 351.000000 351.000000 351.000000 mean 0.891738 0.0 0.641342 ... -0.003794 0.349364 0.014480 std 0.311155 0.0 0.497708 ... 0.513574 0.522663 0.468337 min 0.000000 0.0 -1.000000 ... -1.000000 -1.000000 -1.000000 25% 1.000000 0.0 0.472135 ... -0.242595 0.000000 -0.165350 50% 1.000000 0.0 0.871110 ... 0.000000 0.409560 0.000000 75% 1.000000 0.0 1.000000 ... 0.200120 0.813765 0.171660 max 1.000000 0.0 1.000000 ... 1.000000 1.000000 1.000000 |
A histogram plot is then created for each variable.
We can see that many variables have a Gaussian or Gaussian-like distribution.
We may have some benefit in using a power transform on each variable in order to make the probability distribution less skewed which will likely improve model performance.

Histograms of the Ionosphere Classification Dataset
Now that we are familiar with the dataset, let’s explore how we might develop a neural network model.
Neural Network Learning Dynamics
We will develop a Multilayer Perceptron (MLP) model for the dataset using TensorFlow.
We cannot know what model architecture of learning hyperparameters would be good or best for this dataset, so we must experiment and discover what works well.
Given that the dataset is small, a small batch size is probably a good idea, e.g. 16 or 32 rows. Using the Adam version of stochastic gradient descent is a good idea when getting started as it will automatically adapts the learning rate and works well on most datasets.
Before we evaluate models in earnest, it is a good idea to review the learning dynamics and tune the model architecture and learning configuration until we have stable learning dynamics, then look at getting the most out of the model.
We can do this by using a simple train/test split of the data and review plots of the learning curves. This will help us see if we are over-learning or under-learning; then we can adapt the configuration accordingly.
First, we must ensure all input variables are floating-point values and encode the target label as integer values 0 and 1.
|
1 2 3 4 5 |
... # ensure all data are floating point values X = X.astype('float32') # encode strings to integer y = LabelEncoder().fit_transform(y) |
Next, we can split the dataset into input and output variables, then into 67/33 train and test sets.
|
1 2 3 4 5 |
... # split into input and output columns X, y = df.values[:, :-1], df.values[:, -1] # split into train and test datasets X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33) |
We can define a minimal MLP model. In this case, we will use one hidden layer with 10 nodes and one output layer (chosen arbitrarily). We will use the ReLU activation function in the hidden layer and the “he_normal” weight initialization, as together, they are a good practice.
The output of the model is a sigmoid activation for binary classification and we will minimize binary cross-entropy loss.
|
1 2 3 4 5 6 7 8 9 |
... # determine the number of input features n_features = X.shape[1] # define model model = Sequential() model.add(Dense(10, activation='relu', kernel_initializer='he_normal', input_shape=(n_features,))) model.add(Dense(1, activation='sigmoid')) # compile the model model.compile(optimizer='adam', loss='binary_crossentropy') |
We will fit the model for 200 training epochs (chosen arbitrarily) with a batch size of 32 because it is a small dataset.
We are fitting the model on raw data, which we think might be a good idea, but it is an important starting point.
|
1 2 3 |
... # fit the model history = model.fit(X_train, y_train, epochs=200, batch_size=32, verbose=0, validation_data=(X_test,y_test)) |
At the end of training, we will evaluate the model’s performance on the test dataset and report performance as the classification accuracy.
|
1 2 3 4 5 6 |
... # predict test set yhat = model.predict_classes(X_test) # evaluate predictions score = accuracy_score(y_test, yhat) print('Accuracy: %.3f' % score) |
Finally, we will plot learning curves of the cross-entropy loss on the train and test sets during training.
|
1 2 3 4 5 6 7 8 9 |
... # plot learning curves pyplot.title('Learning Curves') pyplot.xlabel('Epoch') pyplot.ylabel('Cross Entropy') pyplot.plot(history.history['loss'], label='train') pyplot.plot(history.history['val_loss'], label='val') pyplot.legend() pyplot.show() |
Tying this all together, the complete example of evaluating our first MLP on the ionosphere dataset is listed below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 |
# fit a simple mlp model on the ionosphere and review learning curves from pandas import read_csv from sklearn.model_selection import train_test_split from sklearn.preprocessing import LabelEncoder from sklearn.metrics import accuracy_score from tensorflow.keras import Sequential from tensorflow.keras.layers import Dense from matplotlib import pyplot # load the dataset path = 'https://raw.githubusercontent.com/jbrownlee/Datasets/master/ionosphere.csv' df = read_csv(path, header=None) # split into input and output columns X, y = df.values[:, :-1], df.values[:, -1] # ensure all data are floating point values X = X.astype('float32') # encode strings to integer y = LabelEncoder().fit_transform(y) # split into train and test datasets X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33) # determine the number of input features n_features = X.shape[1] # define model model = Sequential() model.add(Dense(10, activation='relu', kernel_initializer='he_normal', input_shape=(n_features,))) model.add(Dense(1, activation='sigmoid')) # compile the model model.compile(optimizer='adam', loss='binary_crossentropy') # fit the model history = model.fit(X_train, y_train, epochs=200, batch_size=32, verbose=0, validation_data=(X_test,y_test)) # predict test set yhat = model.predict_classes(X_test) # evaluate predictions score = accuracy_score(y_test, yhat) print('Accuracy: %.3f' % score) # plot learning curves pyplot.title('Learning Curves') pyplot.xlabel('Epoch') pyplot.ylabel('Cross Entropy') pyplot.plot(history.history['loss'], label='train') pyplot.plot(history.history['val_loss'], label='val') pyplot.legend() pyplot.show() |
Running the example first fits the model on the training dataset, then reports the classification accuracy on the test dataset.
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
In this case, we can see that the model achieved an accuracy of about 88 percent, which is a good baseline in performance that we might be able to improve upon.
|
1 |
Accuracy: 0.888 |
Line plots of the loss on the train and test sets are then created.
We can see that the model appears to converge but has overfit the training dataset.
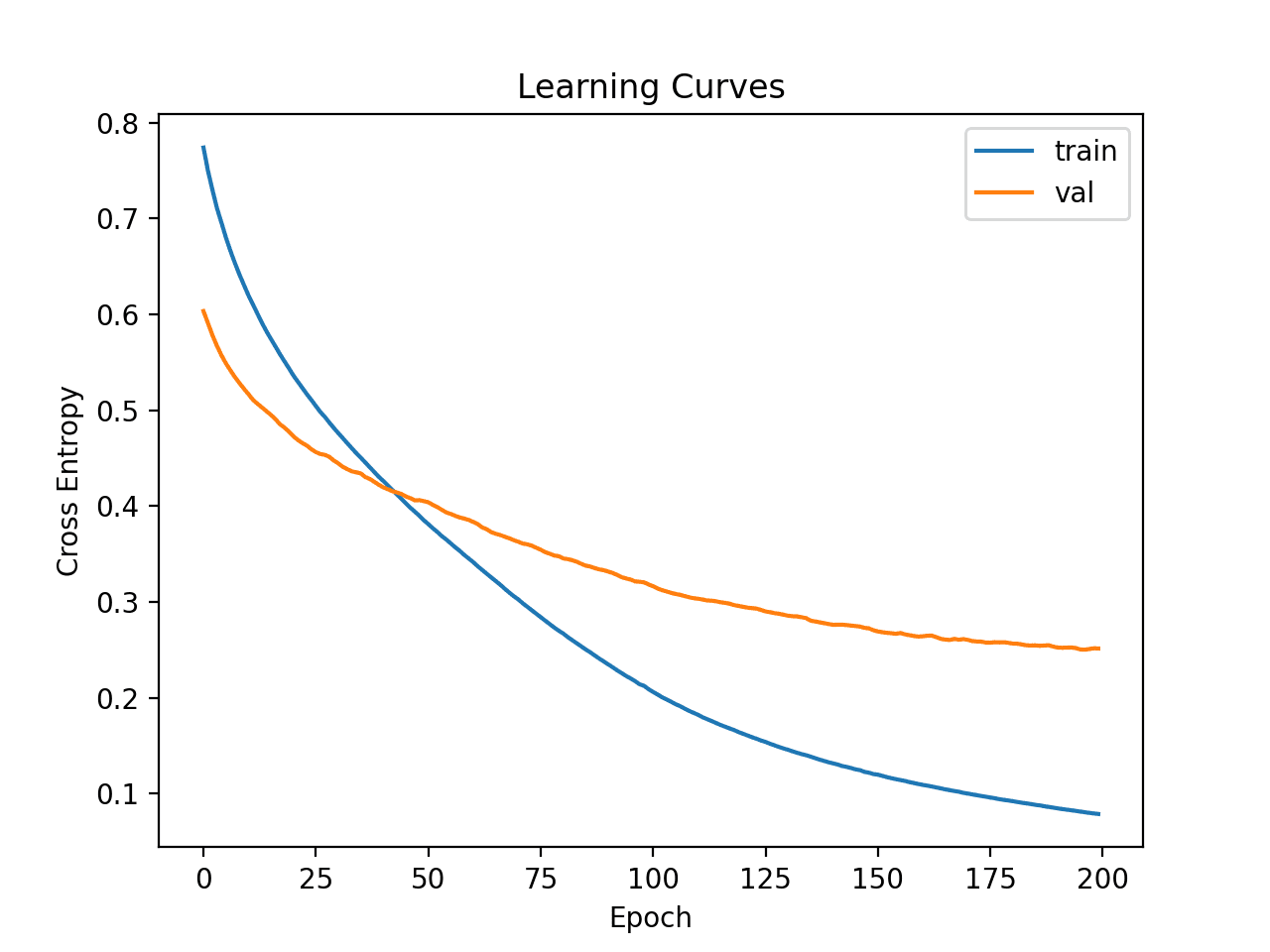
Learning Curves of Simple MLP on Ionosphere Dataset
Let’s try increasing the capacity of the model.
This will slow down learning for the same learning hyperparameters and may offer better accuracy.
We will add a second hidden layer with eight nodes, chosen arbitrarily.
|
1 2 3 4 5 6 |
... # define model model = Sequential() model.add(Dense(10, activation='relu', kernel_initializer='he_normal', input_shape=(n_features,))) model.add(Dense(8, activation='relu', kernel_initializer='he_normal')) model.add(Dense(1, activation='sigmoid')) |
The complete example is listed below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 |
# fit a deeper mlp model on the ionosphere and review learning curves from pandas import read_csv from sklearn.model_selection import train_test_split from sklearn.preprocessing import LabelEncoder from sklearn.metrics import accuracy_score from tensorflow.keras import Sequential from tensorflow.keras.layers import Dense from matplotlib import pyplot # load the dataset path = 'https://raw.githubusercontent.com/jbrownlee/Datasets/master/ionosphere.csv' df = read_csv(path, header=None) # split into input and output columns X, y = df.values[:, :-1], df.values[:, -1] # ensure all data are floating point values X = X.astype('float32') # encode strings to integer y = LabelEncoder().fit_transform(y) # split into train and test datasets X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33) # determine the number of input features n_features = X.shape[1] # define model model = Sequential() model.add(Dense(10, activation='relu', kernel_initializer='he_normal', input_shape=(n_features,))) model.add(Dense(8, activation='relu', kernel_initializer='he_normal')) model.add(Dense(1, activation='sigmoid')) # compile the model model.compile(optimizer='adam', loss='binary_crossentropy') # fit the model history = model.fit(X_train, y_train, epochs=200, batch_size=32, verbose=0, validation_data=(X_test,y_test)) # predict test set yhat = model.predict_classes(X_test) # evaluate predictions score = accuracy_score(y_test, yhat) print('Accuracy: %.3f' % score) # plot learning curves pyplot.title('Learning Curves') pyplot.xlabel('Epoch') pyplot.ylabel('Cross Entropy') pyplot.plot(history.history['loss'], label='train') pyplot.plot(history.history['val_loss'], label='val') pyplot.legend() |
Running the example first fits the model on the training dataset, then reports the accuracy on the test dataset.
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
In this case, we can see a slight improvement in accuracy to about 93 percent, although the high variance of the train/test split means that this evaluation is not reliable.
|
1 |
Accuracy: 0.931 |
Learning curves for the loss on the train and test sets are then plotted. We can see that the model still appears to show an overfitting behavior.
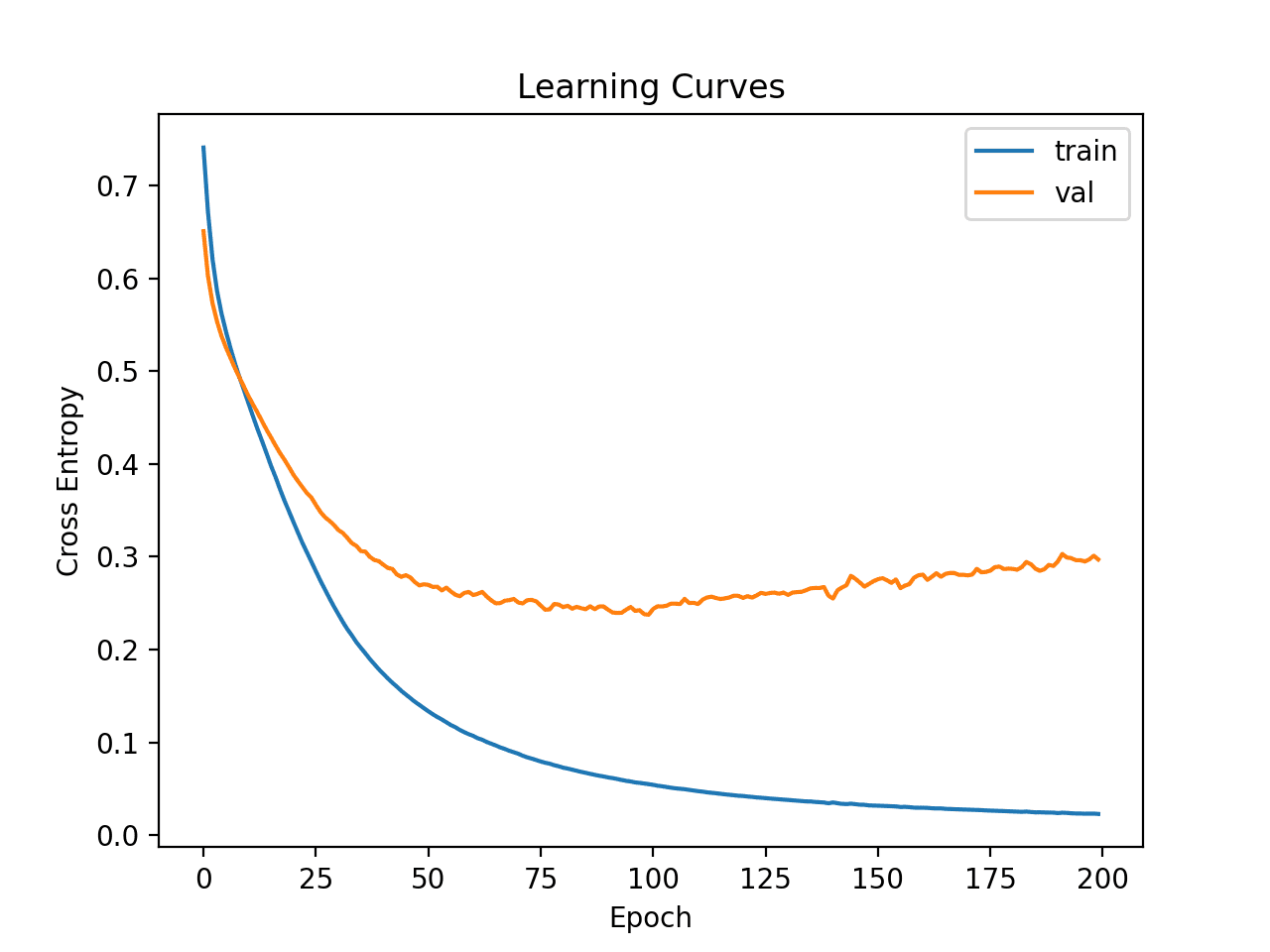
Learning Curves of Deeper MLP on the Ionosphere Dataset
Finally, we can try a wider network.
We will increase the number of nodes in the first hidden layer from 10 to 50, and in the second hidden layer from 8 to 10.
This will add more capacity to the model, slow down learning, and may further improve results.
|
1 2 3 4 5 6 |
... # define model model = Sequential() model.add(Dense(50, activation='relu', kernel_initializer='he_normal', input_shape=(n_features,))) model.add(Dense(10, activation='relu', kernel_initializer='he_normal')) model.add(Dense(1, activation='sigmoid')) |
We will also reduce the number of training epochs from 200 to 100.
|
1 2 3 |
... # fit the model history = model.fit(X_train, y_train, epochs=100, batch_size=32, verbose=0, validation_data=(X_test,y_test)) |
The complete example is listed below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 |
# fit a wider mlp model on the ionosphere and review learning curves from pandas import read_csv from sklearn.model_selection import train_test_split from sklearn.preprocessing import LabelEncoder from sklearn.metrics import accuracy_score from tensorflow.keras import Sequential from tensorflow.keras.layers import Dense from matplotlib import pyplot # load the dataset path = 'https://raw.githubusercontent.com/jbrownlee/Datasets/master/ionosphere.csv' df = read_csv(path, header=None) # split into input and output columns X, y = df.values[:, :-1], df.values[:, -1] # ensure all data are floating point values X = X.astype('float32') # encode strings to integer y = LabelEncoder().fit_transform(y) # split into train and test datasets X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33) # determine the number of input features n_features = X.shape[1] # define model model = Sequential() model.add(Dense(50, activation='relu', kernel_initializer='he_normal', input_shape=(n_features,))) model.add(Dense(10, activation='relu', kernel_initializer='he_normal')) model.add(Dense(1, activation='sigmoid')) # compile the model model.compile(optimizer='adam', loss='binary_crossentropy') # fit the model history = model.fit(X_train, y_train, epochs=100, batch_size=32, verbose=0, validation_data=(X_test,y_test)) # predict test set yhat = model.predict_classes(X_test) # evaluate predictions score = accuracy_score(y_test, yhat) print('Accuracy: %.3f' % score) # plot learning curves pyplot.title('Learning Curves') pyplot.xlabel('Epoch') pyplot.ylabel('Cross Entropy') pyplot.plot(history.history['loss'], label='train') pyplot.plot(history.history['val_loss'], label='val') pyplot.legend() pyplot.show() |
Running the example first fits the model on the training dataset, then reports the accuracy on the test dataset.
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
In this case, the model achieves a better accuracy score, with a value of about 94 percent. We will ignore model performance for now.
|
1 |
Accuracy: 0.940 |
Line plots of the learning curves are created showing that the model achieved a reasonable fit and had more than enough time to converge.
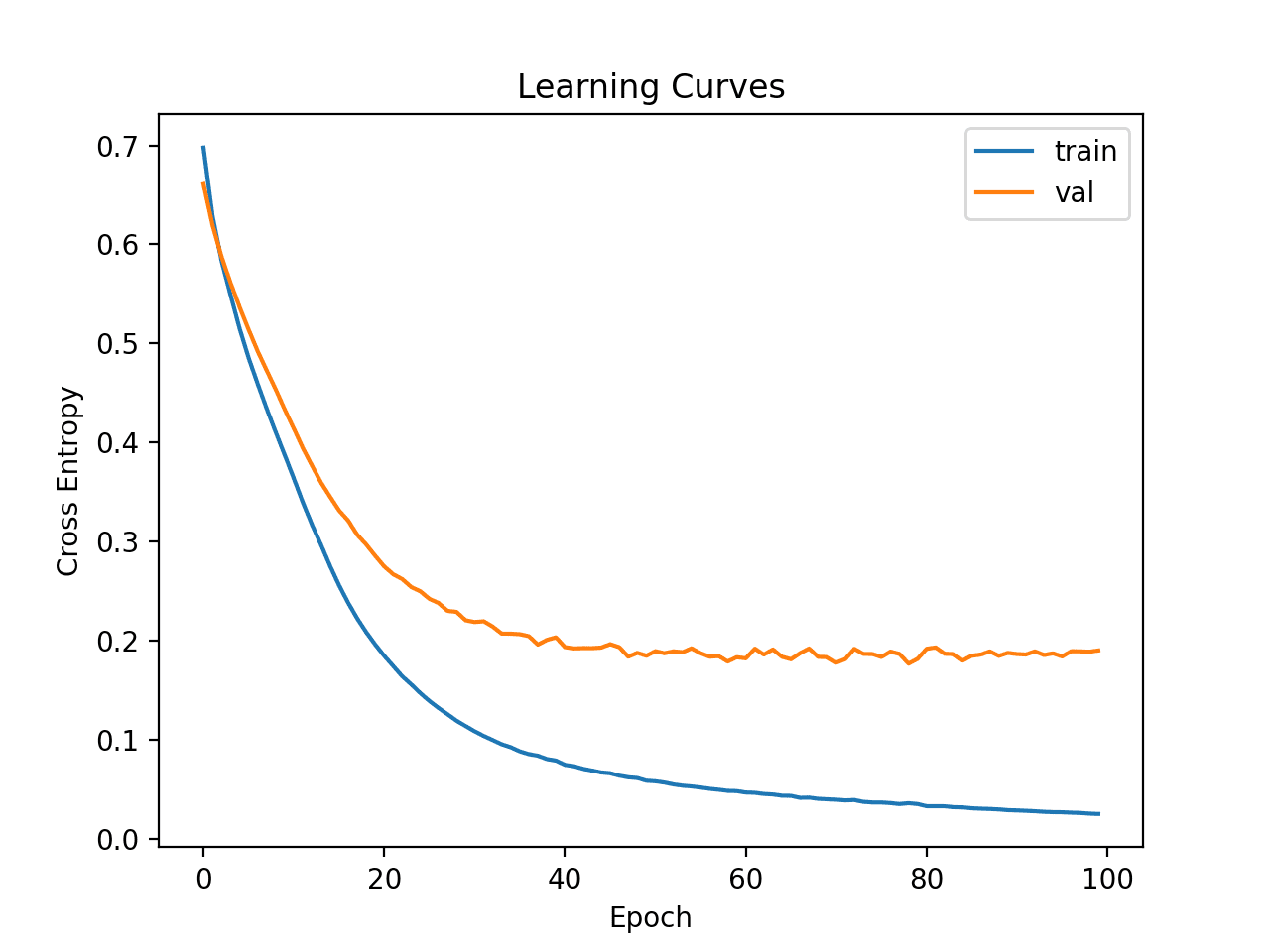
Learning Curves of Wider MLP on the Ionosphere Dataset
Now that we have some idea of the learning dynamics for simple MLP models on the dataset, we can look at evaluating the performance of the models as well as tuning the configuration of the models.
Evaluating and Tuning MLP Models
The k-fold cross-validation procedure can provide a more reliable estimate of MLP performance, although it can be very slow.
This is because k models must be fit and evaluated. This is not a problem when the dataset size is small, such as the ionosphere dataset.
We can use the StratifiedKFold class and enumerate each fold manually, fit the model, evaluate it, and then report the mean of the evaluation scores at the end of the procedure.
|
1 2 3 4 5 6 7 8 9 10 |
# prepare cross validation kfold = KFold(10) # enumerate splits scores = list() for train_ix, test_ix in kfold.split(X, y): # fit and evaluate the model... ... ... # summarize all scores print('Mean Accuracy: %.3f (%.3f)' % (mean(scores), std(scores))) |
We can use this framework to develop a reliable estimate of MLP model performance with a range of different data preparations, model architectures, and learning configurations.
It is important that we first developed an understanding of the learning dynamics of the model on the dataset in the previous section before using k-fold cross-validation to estimate the performance. If we started to tune the model directly, we might get good results, but if not, we might have no idea of why, e.g. that the model was over or under fitting.
If we make large changes to the model again, it is a good idea to go back and confirm that the model is converging appropriately.
The complete example of this framework to evaluate the base MLP model from the previous section is listed below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 |
# k-fold cross-validation of base model for the ionosphere dataset from numpy import mean from numpy import std from pandas import read_csv from sklearn.model_selection import StratifiedKFold from sklearn.preprocessing import LabelEncoder from sklearn.metrics import accuracy_score from tensorflow.keras import Sequential from tensorflow.keras.layers import Dense from matplotlib import pyplot # load the dataset path = 'https://raw.githubusercontent.com/jbrownlee/Datasets/master/ionosphere.csv' df = read_csv(path, header=None) # split into input and output columns X, y = df.values[:, :-1], df.values[:, -1] # ensure all data are floating point values X = X.astype('float32') # encode strings to integer y = LabelEncoder().fit_transform(y) # prepare cross validation kfold = StratifiedKFold(10) # enumerate splits scores = list() for train_ix, test_ix in kfold.split(X, y): # split data X_train, X_test, y_train, y_test = X[train_ix], X[test_ix], y[train_ix], y[test_ix] # determine the number of input features n_features = X.shape[1] # define model model = Sequential() model.add(Dense(50, activation='relu', kernel_initializer='he_normal', input_shape=(n_features,))) model.add(Dense(10, activation='relu', kernel_initializer='he_normal')) model.add(Dense(1, activation='sigmoid')) # compile the model model.compile(optimizer='adam', loss='binary_crossentropy') # fit the model model.fit(X_train, y_train, epochs=100, batch_size=32, verbose=0) # predict test set yhat = model.predict_classes(X_test) # evaluate predictions score = accuracy_score(y_test, yhat) print('>%.3f' % score) scores.append(score) # summarize all scores print('Mean Accuracy: %.3f (%.3f)' % (mean(scores), std(scores))) |
Running the example reports the model performance each iteration of the evaluation procedure and reports the mean and standard deviation of classification accuracy at the end of the run.
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
In this case, we can see that the MLP model achieved a mean accuracy of about 93.4 percent.
We will use this result as our baseline to see if we can achieve better performance.
|
1 2 3 4 5 6 7 8 9 10 11 |
>0.972 >0.886 >0.943 >0.886 >0.914 >0.943 >0.943 >1.000 >0.971 >0.886 Mean Accuracy: 0.934 (0.039) |
Next, let’s try adding regularization to reduce overfitting of the model.
In this case, we can add dropout layers between the hidden layers of the network. For example:
|
1 2 3 4 5 6 7 8 |
... # define model model = Sequential() model.add(Dense(50, activation='relu', kernel_initializer='he_normal', input_shape=(n_features,))) model.add(Dropout(0.4)) model.add(Dense(10, activation='relu', kernel_initializer='he_normal')) model.add(Dropout(0.4)) model.add(Dense(1, activation='sigmoid')) |
The complete example of the MLP model with dropout is listed below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 |
# k-fold cross-validation of the MLP with dropout for the ionosphere dataset from numpy import mean from numpy import std from pandas import read_csv from sklearn.model_selection import StratifiedKFold from sklearn.preprocessing import LabelEncoder from sklearn.metrics import accuracy_score from tensorflow.keras import Sequential from tensorflow.keras.layers import Dense from tensorflow.keras.layers import Dropout from matplotlib import pyplot # load the dataset path = 'https://raw.githubusercontent.com/jbrownlee/Datasets/master/ionosphere.csv' df = read_csv(path, header=None) # split into input and output columns X, y = df.values[:, :-1], df.values[:, -1] # ensure all data are floating point values X = X.astype('float32') # encode strings to integer y = LabelEncoder().fit_transform(y) # prepare cross validation kfold = StratifiedKFold(10) # enumerate splits scores = list() for train_ix, test_ix in kfold.split(X, y): # split data X_train, X_test, y_train, y_test = X[train_ix], X[test_ix], y[train_ix], y[test_ix] # determine the number of input features n_features = X.shape[1] # define model model = Sequential() model.add(Dense(50, activation='relu', kernel_initializer='he_normal', input_shape=(n_features,))) model.add(Dropout(0.4)) model.add(Dense(10, activation='relu', kernel_initializer='he_normal')) model.add(Dropout(0.4)) model.add(Dense(1, activation='sigmoid')) # compile the model model.compile(optimizer='adam', loss='binary_crossentropy') # fit the model model.fit(X_train, y_train, epochs=100, batch_size=32, verbose=0) # predict test set yhat = model.predict_classes(X_test) # evaluate predictions score = accuracy_score(y_test, yhat) print('>%.3f' % score) scores.append(score) # summarize all scores print('Mean Accuracy: %.3f (%.3f)' % (mean(scores), std(scores))) |
Running reports the mean and standard deviation of the classification accuracy at the end of the run.
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
In this case, we can see that the MLP model with dropout achieves better results with an accuracy of about 94.6 percent compared to 93.4 percent without dropout
|
1 |
Mean Accuracy: 0.946 (0.043) |
Finally, we will try reducing the batch size from 32 down to 8.
This will result in more noisy gradients and may also slow down the speed at which the model is learning the problem.
|
1 2 3 |
... # fit the model model.fit(X_train, y_train, epochs=100, batch_size=8, verbose=0) |
The complete example is listed below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 |
# k-fold cross-validation of the MLP with dropout for the ionosphere dataset from numpy import mean from numpy import std from pandas import read_csv from sklearn.model_selection import StratifiedKFold from sklearn.preprocessing import LabelEncoder from sklearn.metrics import accuracy_score from tensorflow.keras import Sequential from tensorflow.keras.layers import Dense from tensorflow.keras.layers import Dropout from matplotlib import pyplot # load the dataset path = 'https://raw.githubusercontent.com/jbrownlee/Datasets/master/ionosphere.csv' df = read_csv(path, header=None) # split into input and output columns X, y = df.values[:, :-1], df.values[:, -1] # ensure all data are floating point values X = X.astype('float32') # encode strings to integer y = LabelEncoder().fit_transform(y) # prepare cross validation kfold = StratifiedKFold(10) # enumerate splits scores = list() for train_ix, test_ix in kfold.split(X, y): # split data X_train, X_test, y_train, y_test = X[train_ix], X[test_ix], y[train_ix], y[test_ix] # determine the number of input features n_features = X.shape[1] # define model model = Sequential() model.add(Dense(50, activation='relu', kernel_initializer='he_normal', input_shape=(n_features,))) model.add(Dropout(0.4)) model.add(Dense(10, activation='relu', kernel_initializer='he_normal')) model.add(Dropout(0.4)) model.add(Dense(1, activation='sigmoid')) # compile the model model.compile(optimizer='adam', loss='binary_crossentropy') # fit the model model.fit(X_train, y_train, epochs=100, batch_size=8, verbose=0) # predict test set yhat = model.predict_classes(X_test) # evaluate predictions score = accuracy_score(y_test, yhat) print('>%.3f' % score) scores.append(score) # summarize all scores print('Mean Accuracy: %.3f (%.3f)' % (mean(scores), std(scores))) |
Running reports the mean and standard deviation of the classification accuracy at the end of the run.
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
In this case, we can see that the MLP model with dropout achieves slightly better results with an accuracy of about 94.9 percent.
|
1 |
Mean Accuracy: 0.949 (0.042) |
We will use this configuration as our final model.
We could continue to test alternate configurations to the model architecture (more or fewer nodes or layers), learning hyperparameters (more or fewer batches), and data transforms.
I leave this as an exercise; let me know what you discover. Can you get better results?
Post your results in the comments below, I’d love to see what you get.
Next, let’s look at how we might fit a final model and use it to make predictions.
Final Model and Make Predictions
Once we choose a model configuration, we can train a final model on all available data and use it to make predictions on new data.
In this case, we will use the model with dropout and a small batch size as our final model.
We can prepare the data and fit the model as before, although on the entire dataset instead of a training subset of the dataset.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 |
... # split into input and output columns X, y = df.values[:, :-1], df.values[:, -1] # ensure all data are floating point values X = X.astype('float32') # encode strings to integer le = LabelEncoder() y = le.fit_transform(y) # determine the number of input features n_features = X.shape[1] # define model model = Sequential() model.add(Dense(50, activation='relu', kernel_initializer='he_normal', input_shape=(n_features,))) model.add(Dropout(0.4)) model.add(Dense(10, activation='relu', kernel_initializer='he_normal')) model.add(Dropout(0.4)) model.add(Dense(1, activation='sigmoid')) # compile the model model.compile(optimizer='adam', loss='binary_crossentropy') |
We can then use this model to make predictions on new data.
First, we can define a row of new data.
|
1 2 3 |
... # define a row of new data row = [1,0,0.99539,-0.05889,0.85243,0.02306,0.83398,-0.37708,1,0.03760,0.85243,-0.17755,0.59755,-0.44945,0.60536,-0.38223,0.84356,-0.38542,0.58212,-0.32192,0.56971,-0.29674,0.36946,-0.47357,0.56811,-0.51171,0.41078,-0.46168,0.21266,-0.34090,0.42267,-0.54487,0.18641,-0.45300] |
Note: I took this row from the first row of the dataset and the expected label is a ‘g‘.
We can then make a prediction.
|
1 2 3 |
... # make prediction yhat = model.predict_classes([row]) |
Then invert the transform on the prediction, so we can use or interpret the result in the correct label.
|
1 2 3 |
... # invert transform to get label for class yhat = le.inverse_transform(yhat) |
And in this case, we will simply report the prediction.
|
1 2 3 |
... # report prediction print('Predicted: %s' % (yhat[0])) |
Tying this all together, the complete example of fitting a final model for the ionosphere dataset and using it to make a prediction on new data is listed below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 |
# fit a final model and make predictions on new data for the ionosphere dataset from pandas import read_csv from sklearn.preprocessing import LabelEncoder from sklearn.metrics import accuracy_score from tensorflow.keras import Sequential from tensorflow.keras.layers import Dense from tensorflow.keras.layers import Dropout # load the dataset path = 'https://raw.githubusercontent.com/jbrownlee/Datasets/master/ionosphere.csv' df = read_csv(path, header=None) # split into input and output columns X, y = df.values[:, :-1], df.values[:, -1] # ensure all data are floating point values X = X.astype('float32') # encode strings to integer le = LabelEncoder() y = le.fit_transform(y) # determine the number of input features n_features = X.shape[1] # define model model = Sequential() model.add(Dense(50, activation='relu', kernel_initializer='he_normal', input_shape=(n_features,))) model.add(Dropout(0.4)) model.add(Dense(10, activation='relu', kernel_initializer='he_normal')) model.add(Dropout(0.4)) model.add(Dense(1, activation='sigmoid')) # compile the model model.compile(optimizer='adam', loss='binary_crossentropy') # fit the model model.fit(X, y, epochs=100, batch_size=8, verbose=0) # define a row of new data row = [1,0,0.99539,-0.05889,0.85243,0.02306,0.83398,-0.37708,1,0.03760,0.85243,-0.17755,0.59755,-0.44945,0.60536,-0.38223,0.84356,-0.38542,0.58212,-0.32192,0.56971,-0.29674,0.36946,-0.47357,0.56811,-0.51171,0.41078,-0.46168,0.21266,-0.34090,0.42267,-0.54487,0.18641,-0.45300] # make prediction yhat = model.predict_classes([row]) # invert transform to get label for class yhat = le.inverse_transform(yhat) # report prediction print('Predicted: %s' % (yhat[0])) |
Running the example fits the model on the entire dataset and makes a prediction for a single row of new data.
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
In this case, we can see that the model predicted a “g” label for the input row.
|
1 |
Predicted: g |
Further Reading
This section provides more resources on the topic if you are looking to go deeper.
Tutorials
- Best Results for Standard Machine Learning Datasets
- TensorFlow 2 Tutorial: Get Started in Deep Learning With tf.keras
- A Gentle Introduction to k-fold Cross-Validation
Summary
In this tutorial, you discovered how to develop a Multilayer Perceptron neural network model for the ionosphere binary classification dataset.
Specifically, you learned:
- How to load and summarize the ionosphere dataset and use the results to suggest data preparations and model configurations to use.
- How to explore the learning dynamics of simple MLP models on the dataset.
- How to develop robust estimates of model performance, tune model performance and make predictions on new data.
Do you have any questions?
Ask your questions in the comments below and I will do my best to answer.



Wow, excellent tutorial and it’s fresh off the press! I just started DL so perfect timing. There’s a lot to study here so I have my homework cut out for me.
Thank you, Jason.
You’re welcome.
Awesome tutorial.
Thanks for sharing, Jason Brownlee
You’re very welcome.
Awesome ! Good technical stuff. Its really hard to fit vast content. Highly appreciate your effort @Jason Brownlee.
You’re welcome, I’m happy it’s helpful.
Amazing presentation and the way of explaining concepts in layman language is commendable. Kindly tell me how to get access to the collab notebook. I read this type of concept from article of sachin dev duggal which (CEO & Founder of Builder AI).
Thanks.
This can help with colab:
https://machinelearningmastery.com/faq/single-faq/do-code-examples-run-on-google-colab
Great article!! More understandable
Thanks.
sir as i want to apply horizontal ensembling to ionosphere data it gives me error
File “C:\Users\LENOVO\untitled4.py”, line 79, in
members = load_all_models(950, 1000)
File “C:\Users\LENOVO\untitled4.py”, line 38, in load_all_models
model = load_model(filename)
File “C:\Users\LENOVO\anaconda3\envs\tensorflow\lib\site-packages\keras\engine\saving.py”, line 492, in load_wrapper
return load_function(*args, **kwargs)
File “C:\Users\LENOVO\anaconda3\envs\tensorflow\lib\site-packages\keras\engine\saving.py”, line 584, in load_model
model = _deserialize_model(h5dict, custom_objects, compile)
File “C:\Users\LENOVO\anaconda3\envs\tensorflow\lib\site-packages\keras\engine\saving.py”, line 274, in _deserialize_model
model = model_from_config(model_config, custom_objects=custom_objects)
File “C:\Users\LENOVO\anaconda3\envs\tensorflow\lib\site-packages\keras\engine\saving.py”, line 627, in model_from_config
return deserialize(config, custom_objects=custom_objects)
File “C:\Users\LENOVO\anaconda3\envs\tensorflow\lib\site-packages\keras\layers\__init__.py”, line 168, in deserialize
printable_module_name=’layer’)
File “C:\Users\LENOVO\anaconda3\envs\tensorflow\lib\site-packages\keras\utils\generic_utils.py”, line 147, in deserialize_keras_object
list(custom_objects.items())))
File “C:\Users\LENOVO\anaconda3\envs\tensorflow\lib\site-packages\keras\engine\sequential.py”, line 301, in from_config
custom_objects=custom_objects)
File “C:\Users\LENOVO\anaconda3\envs\tensorflow\lib\site-packages\keras\layers\__init__.py”, line 168, in deserialize
printable_module_name=’layer’)
File “C:\Users\LENOVO\anaconda3\envs\tensorflow\lib\site-packages\keras\utils\generic_utils.py”, line 149, in deserialize_keras_object
return cls.from_config(config[‘config’])
File “C:\Users\LENOVO\anaconda3\envs\tensorflow\lib\site-packages\keras\engine\base_layer.py”, line 1179, in from_config
return cls(**config)
File “C:\Users\LENOVO\anaconda3\envs\tensorflow\lib\site-packages\keras\legacy\interfaces.py”, line 91, in wrapper
return func(*args, **kwargs)
File “C:\Users\LENOVO\anaconda3\envs\tensorflow\lib\site-packages\keras\layers\core.py”, line 877, in __init__
self.kernel_initializer = initializers.get(kernel_initializer)
File “C:\Users\LENOVO\anaconda3\envs\tensorflow\lib\site-packages\keras\initializers.py”, line 515, in get
return deserialize(identifier)
File “C:\Users\LENOVO\anaconda3\envs\tensorflow\lib\site-packages\keras\initializers.py”, line 510, in deserialize
printable_module_name=’initializer’)
File “C:\Users\LENOVO\anaconda3\envs\tensorflow\lib\site-packages\keras\utils\generic_utils.py”, line 149, in deserialize_keras_object
return cls.from_config(config[‘config’])
File “C:\Users\LENOVO\anaconda3\envs\tensorflow\lib\site-packages\keras\initializers.py”, line 30, in from_config
return cls(**config)
File “C:\Users\LENOVO\anaconda3\envs\tensorflow\lib\site-packages\keras\initializers.py”, line 204, in __init__
‘but got’, distribution)
ValueError: (‘Invalid
distributionargument: expected one of {“normal”, “uniform”} but got’, ‘truncated_normal’)kindly help me to solve this
Sorry I don’t know the cause of your error.
Perhaps you can post your code, data and error on stackoverflow.com