Machine learning and deep learning models, like those in Keras, require all input and output variables to be numeric.
This means that if your data contains categorical data, you must encode it to numbers before you can fit and evaluate a model.
The two most popular techniques are an integer encoding and a one hot encoding, although a newer technique called learned embedding may provide a useful middle ground between these two methods.
In this tutorial, you will discover how to encode categorical data when developing neural network models in Keras.
After completing this tutorial, you will know:
- The challenge of working with categorical data when using machine learning and deep learning models.
- How to integer encode and one hot encode categorical variables for modeling.
- How to learn an embedding distributed representation as part of a neural network for categorical variables.
Kick-start your project with my new book Deep Learning With Python, including step-by-step tutorials and the Python source code files for all examples.
Let’s get started.

How to Encode Categorical Data for Deep Learning in Keras
Photo by Ken Dixon, some rights reserved.
Tutorial Overview
This tutorial is divided into five parts; they are:
- The Challenge With Categorical Data
- Breast Cancer Categorical Dataset
- How to Ordinal Encode Categorical Data
- How to One Hot Encode Categorical Data
- How to Use a Learned Embedding for Categorical Data
The Challenge With Categorical Data
A categorical variable is a variable whose values take on the value of labels.
For example, the variable may be “color” and may take on the values “red,” “green,” and “blue.”
Sometimes, the categorical data may have an ordered relationship between the categories, such as “first,” “second,” and “third.” This type of categorical data is referred to as ordinal and the additional ordering information can be useful.
Machine learning algorithms and deep learning neural networks require that input and output variables are numbers.
This means that categorical data must be encoded to numbers before we can use it to fit and evaluate a model.
There are many ways to encode categorical variables for modeling, although the three most common are as follows:
- Integer Encoding: Where each unique label is mapped to an integer.
- One Hot Encoding: Where each label is mapped to a binary vector.
- Learned Embedding: Where a distributed representation of the categories is learned.
We will take a closer look at how to encode categorical data for training a deep learning neural network in Keras using each one of these methods.
Breast Cancer Categorical Dataset
As the basis of this tutorial, we will use the so-called “Breast cancer” dataset that has been widely studied in machine learning since the 1980s.
The dataset classifies breast cancer patient data as either a recurrence or no recurrence of cancer. There are 286 examples and nine input variables. It is a binary classification problem.
A reasonable classification accuracy score on this dataset is between 68% and 73%. We will aim for this region, but note that the models in this tutorial are not optimized: they are designed to demonstrate encoding schemes.
You can download the dataset and save the file as “breast-cancer.csv” in your current working directory.
Looking at the data, we can see that all nine input variables are categorical.
Specifically, all variables are quoted strings; some are ordinal and some are not.
|
1 2 3 4 5 6 |
'40-49','premeno','15-19','0-2','yes','3','right','left_up','no','recurrence-events' '50-59','ge40','15-19','0-2','no','1','right','central','no','no-recurrence-events' '50-59','ge40','35-39','0-2','no','2','left','left_low','no','recurrence-events' '40-49','premeno','35-39','0-2','yes','3','right','left_low','yes','no-recurrence-events' '40-49','premeno','30-34','3-5','yes','2','left','right_up','no','recurrence-events' ... |
We can load this dataset into memory using the Pandas library.
|
1 2 3 4 5 |
... # load the dataset as a pandas DataFrame data = read_csv(filename, header=None) # retrieve numpy array dataset = data.values |
Once loaded, we can split the columns into input (X) and output (y) for modeling.
|
1 2 3 4 |
... # split into input (X) and output (y) variables X = dataset[:, :-1] y = dataset[:,-1] |
Finally, we can force all fields in the input data to be string, just in case Pandas tried to map some automatically to numbers (it does try).
We can also reshape the output variable to be one column (e.g. a 2D shape).
|
1 2 3 4 5 |
... # format all fields as string X = X.astype(str) # reshape target to be a 2d array y = y.reshape((len(y), 1)) |
We can tie all of this together into a helpful function that we can reuse later.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 |
# load the dataset def load_dataset(filename): # load the dataset as a pandas DataFrame data = read_csv(filename, header=None) # retrieve numpy array dataset = data.values # split into input (X) and output (y) variables X = dataset[:, :-1] y = dataset[:,-1] # format all fields as string X = X.astype(str) # reshape target to be a 2d array y = y.reshape((len(y), 1)) return X, y |
Once loaded, we can split the data into training and test sets so that we can fit and evaluate a deep learning model.
We will use the train_test_split() function from scikit-learn and use 67% of the data for training and 33% for testing.
|
1 2 3 4 5 |
... # load the dataset X, y = load_dataset('breast-cancer.csv') # split into train and test sets X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=1) |
Tying all of these elements together, the complete example of loading, splitting, and summarizing the raw categorical dataset is listed below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 |
# load and summarize the dataset from pandas import read_csv from sklearn.model_selection import train_test_split # load the dataset def load_dataset(filename): # load the dataset as a pandas DataFrame data = read_csv(filename, header=None) # retrieve numpy array dataset = data.values # split into input (X) and output (y) variables X = dataset[:, :-1] y = dataset[:,-1] # format all fields as string X = X.astype(str) # reshape target to be a 2d array y = y.reshape((len(y), 1)) return X, y # load the dataset X, y = load_dataset('breast-cancer.csv') # split into train and test sets X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=1) # summarize print('Train', X_train.shape, y_train.shape) print('Test', X_test.shape, y_test.shape) |
Running the example reports the size of the input and output elements of the train and test sets.
We can see that we have 191 examples for training and 95 for testing.
|
1 2 |
Train (191, 9) (191, 1) Test (95, 9) (95, 1) |
Now that we are familiar with the dataset, let’s look at how we can encode it for modeling.
How to Ordinal Encode Categorical Data
An ordinal encoding involves mapping each unique label to an integer value.
As such, it is sometimes referred to simply as an integer encoding.
This type of encoding is really only appropriate if there is a known relationship between the categories.
This relationship does exist for some of the variables in the dataset, and ideally, this should be harnessed when preparing the data.
In this case, we will ignore any possible existing ordinal relationship and assume all variables are categorical. It can still be helpful to use an ordinal encoding, at least as a point of reference with other encoding schemes.
We can use the OrdinalEncoder() from scikit-learn to encode each variable to integers. This is a flexible class and does allow the order of the categories to be specified as arguments if any such order is known.
Note: I will leave it as an exercise for you to update the example below to try specifying the order for those variables that have a natural ordering and see if it has an impact on model performance.
The best practice when encoding variables is to fit the encoding on the training dataset, then apply it to the train and test datasets.
The function below, named prepare_inputs(), takes the input data for the train and test sets and encodes it using an ordinal encoding.
|
1 2 3 4 5 6 7 |
# prepare input data def prepare_inputs(X_train, X_test): oe = OrdinalEncoder() oe.fit(X_train) X_train_enc = oe.transform(X_train) X_test_enc = oe.transform(X_test) return X_train_enc, X_test_enc |
We also need to prepare the target variable.
It is a binary classification problem, so we need to map the two class labels to 0 and 1.
This is a type of ordinal encoding, and scikit-learn provides the LabelEncoder class specifically designed for this purpose. We could just as easily use the OrdinalEncoder and achieve the same result, although the LabelEncoder is designed for encoding a single variable.
The prepare_targets() integer encodes the output data for the train and test sets.
|
1 2 3 4 5 6 7 |
# prepare target def prepare_targets(y_train, y_test): le = LabelEncoder() le.fit(y_train) y_train_enc = le.transform(y_train) y_test_enc = le.transform(y_test) return y_train_enc, y_test_enc |
We can call these functions to prepare our data.
|
1 2 3 4 5 |
... # prepare input data X_train_enc, X_test_enc = prepare_inputs(X_train, X_test) # prepare output data y_train_enc, y_test_enc = prepare_targets(y_train, y_test) |
We can now define a neural network model.
We will use the same general model in all of these examples. Specifically, a MultiLayer Perceptron (MLP) neural network with one hidden layer with 10 nodes, and one node in the output layer for making binary classifications.
Without going into too much detail, the code below defines the model, fits it on the training dataset, and then evaluates it on the test dataset.
|
1 2 3 4 5 6 7 8 9 10 11 12 |
... # define the model model = Sequential() model.add(Dense(10, input_dim=X_train_enc.shape[1], activation='relu', kernel_initializer='he_normal')) model.add(Dense(1, activation='sigmoid')) # compile the keras model model.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy']) # fit the keras model on the dataset model.fit(X_train_enc, y_train_enc, epochs=100, batch_size=16, verbose=2) # evaluate the keras model _, accuracy = model.evaluate(X_test_enc, y_test_enc, verbose=0) print('Accuracy: %.2f' % (accuracy*100)) |
If you are new to developing neural networks in Keras, I recommend this tutorial:
Tying all of this together, the complete example of preparing the data with an ordinal encoding and fitting and evaluating a neural network on the data is listed below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 |
# example of ordinal encoding for a neural network from pandas import read_csv from sklearn.model_selection import train_test_split from sklearn.preprocessing import LabelEncoder from sklearn.preprocessing import OrdinalEncoder from keras.models import Sequential from keras.layers import Dense # load the dataset def load_dataset(filename): # load the dataset as a pandas DataFrame data = read_csv(filename, header=None) # retrieve numpy array dataset = data.values # split into input (X) and output (y) variables X = dataset[:, :-1] y = dataset[:,-1] # format all fields as string X = X.astype(str) # reshape target to be a 2d array y = y.reshape((len(y), 1)) return X, y # prepare input data def prepare_inputs(X_train, X_test): oe = OrdinalEncoder() oe.fit(X_train) X_train_enc = oe.transform(X_train) X_test_enc = oe.transform(X_test) return X_train_enc, X_test_enc # prepare target def prepare_targets(y_train, y_test): le = LabelEncoder() le.fit(y_train) y_train_enc = le.transform(y_train) y_test_enc = le.transform(y_test) return y_train_enc, y_test_enc # load the dataset X, y = load_dataset('breast-cancer.csv') # split into train and test sets X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=1) # prepare input data X_train_enc, X_test_enc = prepare_inputs(X_train, X_test) # prepare output data y_train_enc, y_test_enc = prepare_targets(y_train, y_test) # define the model model = Sequential() model.add(Dense(10, input_dim=X_train_enc.shape[1], activation='relu', kernel_initializer='he_normal')) model.add(Dense(1, activation='sigmoid')) # compile the keras model model.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy']) # fit the keras model on the dataset model.fit(X_train_enc, y_train_enc, epochs=100, batch_size=16, verbose=2) # evaluate the keras model _, accuracy = model.evaluate(X_test_enc, y_test_enc, verbose=0) print('Accuracy: %.2f' % (accuracy*100)) |
Running the example will fit the model in just a few seconds on any modern hardware (no GPU required).
The loss and the accuracy of the model are reported at the end of each training epoch, and finally, the accuracy of the model on the test dataset is reported.
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
In this case, we can see that the model achieved an accuracy of about 70% on the test dataset.
Not bad, given that an ordinal relationship only exists for some of the input variables, and for those where it does, it was not honored in the encoding.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 |
... Epoch 95/100 - 0s - loss: 0.5349 - acc: 0.7696 Epoch 96/100 - 0s - loss: 0.5330 - acc: 0.7539 Epoch 97/100 - 0s - loss: 0.5316 - acc: 0.7592 Epoch 98/100 - 0s - loss: 0.5302 - acc: 0.7696 Epoch 99/100 - 0s - loss: 0.5291 - acc: 0.7644 Epoch 100/100 - 0s - loss: 0.5277 - acc: 0.7644 Accuracy: 70.53 |
This provides a good starting point when working with categorical data.
A better and more general approach is to use a one hot encoding.
How to One Hot Encode Categorical Data
A one hot encoding is appropriate for categorical data where no relationship exists between categories.
It involves representing each categorical variable with a binary vector that has one element for each unique label and marking the class label with a 1 and all other elements 0.
For example, if our variable was “color” and the labels were “red,” “green,” and “blue,” we would encode each of these labels as a three-element binary vector as follows:
- Red: [1, 0, 0]
- Green: [0, 1, 0]
- Blue: [0, 0, 1]
Then each label in the dataset would be replaced with a vector (one column becomes three). This is done for all categorical variables so that our nine input variables or columns become 43 in the case of the breast cancer dataset.
The scikit-learn library provides the OneHotEncoder to automatically one hot encode one or more variables.
The prepare_inputs() function below provides a drop-in replacement function for the example in the previous section. Instead of using an OrdinalEncoder, it uses a OneHotEncoder.
|
1 2 3 4 5 6 7 |
# prepare input data def prepare_inputs(X_train, X_test): ohe = OneHotEncoder() ohe.fit(X_train) X_train_enc = ohe.transform(X_train) X_test_enc = ohe.transform(X_test) return X_train_enc, X_test_enc |
Tying this together, the complete example of one hot encoding the breast cancer categorical dataset and modeling it with a neural network is listed below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 |
# example of one hot encoding for a neural network from pandas import read_csv from sklearn.model_selection import train_test_split from sklearn.preprocessing import LabelEncoder from sklearn.preprocessing import OneHotEncoder from keras.models import Sequential from keras.layers import Dense # load the dataset def load_dataset(filename): # load the dataset as a pandas DataFrame data = read_csv(filename, header=None) # retrieve numpy array dataset = data.values # split into input (X) and output (y) variables X = dataset[:, :-1] y = dataset[:,-1] # format all fields as string X = X.astype(str) # reshape target to be a 2d array y = y.reshape((len(y), 1)) return X, y # prepare input data def prepare_inputs(X_train, X_test): ohe = OneHotEncoder() ohe.fit(X_train) X_train_enc = ohe.transform(X_train) X_test_enc = ohe.transform(X_test) return X_train_enc, X_test_enc # prepare target def prepare_targets(y_train, y_test): le = LabelEncoder() le.fit(y_train) y_train_enc = le.transform(y_train) y_test_enc = le.transform(y_test) return y_train_enc, y_test_enc # load the dataset X, y = load_dataset('breast-cancer.csv') # split into train and test sets X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=1) # prepare input data X_train_enc, X_test_enc = prepare_inputs(X_train, X_test) # prepare output data y_train_enc, y_test_enc = prepare_targets(y_train, y_test) # define the model model = Sequential() model.add(Dense(10, input_dim=X_train_enc.shape[1], activation='relu', kernel_initializer='he_normal')) model.add(Dense(1, activation='sigmoid')) # compile the keras model model.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy']) # fit the keras model on the dataset model.fit(X_train_enc, y_train_enc, epochs=100, batch_size=16, verbose=2) # evaluate the keras model _, accuracy = model.evaluate(X_test_enc, y_test_enc, verbose=0) print('Accuracy: %.2f' % (accuracy*100)) |
The example one hot encodes the input categorical data, and also label encodes the target variable as we did in the previous section. The same neural network model is then fit on the prepared dataset.
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
In this case, the model performs reasonably well, achieving an accuracy of about 72%, close to what was seen in the previous section.
A more fair comparison would be to run each configuration 10 or 30 times and compare performance using the mean accuracy. Recall, that we are more focused on how to encode categorical data in this tutorial rather than getting the best score on this specific dataset.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 |
... Epoch 95/100 - 0s - loss: 0.3837 - acc: 0.8272 Epoch 96/100 - 0s - loss: 0.3823 - acc: 0.8325 Epoch 97/100 - 0s - loss: 0.3814 - acc: 0.8325 Epoch 98/100 - 0s - loss: 0.3795 - acc: 0.8325 Epoch 99/100 - 0s - loss: 0.3788 - acc: 0.8325 Epoch 100/100 - 0s - loss: 0.3773 - acc: 0.8325 Accuracy: 72.63 |
Ordinal and one hot encoding are perhaps the two most popular methods.
A newer technique is similar to one hot encoding and was designed for use with neural networks, called a learned embedding.
How to Use a Learned Embedding for Categorical Data
A learned embedding, or simply an “embedding,” is a distributed representation for categorical data.
Each category is mapped to a distinct vector, and the properties of the vector are adapted or learned while training a neural network. The vector space provides a projection of the categories, allowing those categories that are close or related to cluster together naturally.
This provides both the benefits of an ordinal relationship by allowing any such relationships to be learned from data, and a one hot encoding in providing a vector representation for each category. Unlike one hot encoding, the input vectors are not sparse (do not have lots of zeros). The downside is that it requires learning as part of the model and the creation of many more input variables (columns).
The technique was originally developed to provide a distributed representation for words, e.g. allowing similar words to have similar vector representations. As such, the technique is often referred to as a word embedding, and in the case of text data, algorithms have been developed to learn a representation independent of a neural network. For more on this topic, see the post:
An additional benefit of using an embedding is that the learned vectors that each category is mapped to can be fit in a model that has modest skill, but the vectors can be extracted and used generally as input for the category on a range of different models and applications. That is, they can be learned and reused.
Embeddings can be used in Keras via the Embedding layer.
For an example of learning word embeddings for text data in Keras, see the post:
One embedding layer is required for each categorical variable, and the embedding expects the categories to be ordinal encoded, although no relationship between the categories is assumed.
Each embedding also requires the number of dimensions to use for the distributed representation (vector space). It is common in natural language applications to use 50, 100, or 300 dimensions. For our small example, we will fix the number of dimensions at 10, but this is arbitrary; you should experimenter with other values.
First, we can prepare the input data using an ordinal encoding.
The model we will develop will have one separate embedding for each input variable. Therefore, the model will take nine different input datasets. As such, we will split the input variables and ordinal encode (integer encoding) each separately using the LabelEncoder and return a list of separate prepared train and test input datasets.
The prepare_inputs() function below implements this, enumerating over each input variable, integer encoding each correctly using best practices, and returning lists of encoded train and test variables (or one-variable datasets) that can be used as input for our model later.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 |
# prepare input data def prepare_inputs(X_train, X_test): X_train_enc, X_test_enc = list(), list() # label encode each column for i in range(X_train.shape[1]): le = LabelEncoder() le.fit(X_train[:, i]) # encode train_enc = le.transform(X_train[:, i]) test_enc = le.transform(X_test[:, i]) # store X_train_enc.append(train_enc) X_test_enc.append(test_enc) return X_train_enc, X_test_enc |
Now we can construct the model.
We must construct the model differently in this case because we will have nine input layers, with nine embeddings the outputs of which (the nine different 10-element vectors) need to be concatenated into one long vector before being passed as input to the dense layers.
We can achieve this using the functional Keras API. If you are new to the Keras functional API, see the post:
First, we can enumerate each variable and construct an input layer and connect it to an embedding layer, and store both layers in lists. We need a reference to all of the input layers when defining the model, and we need a reference to each embedding layer to concentrate them with a merge layer.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 |
... # prepare each input head in_layers = list() em_layers = list() for i in range(len(X_train_enc)): # calculate the number of unique inputs n_labels = len(unique(X_train_enc[i])) # define input layer in_layer = Input(shape=(1,)) # define embedding layer em_layer = Embedding(n_labels, 10)(in_layer) # store layers in_layers.append(in_layer) em_layers.append(em_layer) |
We can then merge all of the embedding layers, define the hidden layer and output layer, then define the model.
|
1 2 3 4 5 6 |
... # concat all embeddings merge = concatenate(em_layers) dense = Dense(10, activation='relu', kernel_initializer='he_normal')(merge) output = Dense(1, activation='sigmoid')(dense) model = Model(inputs=in_layers, outputs=output) |
When using a model with multiple inputs, we will need to specify a list that has one dataset for each input, e.g. a list of nine arrays each with one column in the case of our dataset. Thankfully, this is the format we returned from our prepare_inputs() function.
Therefore, fitting and evaluating the model looks like it does in the previous section.
Additionally, we will plot the model by calling the plot_model() function and save it to file. This requires that pygraphviz and pydot are installed, which can be a pain on some systems. If you have trouble, just comment out the import statement and call to plot_model().
|
1 2 3 4 5 6 7 8 9 10 |
... # compile the keras model model.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy']) # plot graph plot_model(model, show_shapes=True, to_file='embeddings.png') # fit the keras model on the dataset model.fit(X_train_enc, y_train_enc, epochs=20, batch_size=16, verbose=2) # evaluate the keras model _, accuracy = model.evaluate(X_test_enc, y_test_enc, verbose=0) print('Accuracy: %.2f' % (accuracy*100)) |
Tying this all together, the complete example of using a separate embedding for each categorical input variable in a multi-input layer model is listed below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 |
# example of learned embedding encoding for a neural network from numpy import unique from pandas import read_csv from sklearn.model_selection import train_test_split from sklearn.preprocessing import LabelEncoder from keras.models import Model from keras.layers import Input from keras.layers import Dense from keras.layers import Embedding from keras.layers.merge import concatenate from keras.utils import plot_model # load the dataset def load_dataset(filename): # load the dataset as a pandas DataFrame data = read_csv(filename, header=None) # retrieve numpy array dataset = data.values # split into input (X) and output (y) variables X = dataset[:, :-1] y = dataset[:,-1] # format all fields as string X = X.astype(str) # reshape target to be a 2d array y = y.reshape((len(y), 1)) return X, y # prepare input data def prepare_inputs(X_train, X_test): X_train_enc, X_test_enc = list(), list() # label encode each column for i in range(X_train.shape[1]): le = LabelEncoder() le.fit(X_train[:, i]) # encode train_enc = le.transform(X_train[:, i]) test_enc = le.transform(X_test[:, i]) # store X_train_enc.append(train_enc) X_test_enc.append(test_enc) return X_train_enc, X_test_enc # prepare target def prepare_targets(y_train, y_test): le = LabelEncoder() le.fit(y_train) y_train_enc = le.transform(y_train) y_test_enc = le.transform(y_test) return y_train_enc, y_test_enc # load the dataset X, y = load_dataset('breast-cancer.csv') # split into train and test sets X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=1) # prepare input data X_train_enc, X_test_enc = prepare_inputs(X_train, X_test) # prepare output data y_train_enc, y_test_enc = prepare_targets(y_train, y_test) # make output 3d y_train_enc = y_train_enc.reshape((len(y_train_enc), 1, 1)) y_test_enc = y_test_enc.reshape((len(y_test_enc), 1, 1)) # prepare each input head in_layers = list() em_layers = list() for i in range(len(X_train_enc)): # calculate the number of unique inputs n_labels = len(unique(X_train_enc[i])) # define input layer in_layer = Input(shape=(1,)) # define embedding layer em_layer = Embedding(n_labels, 10)(in_layer) # store layers in_layers.append(in_layer) em_layers.append(em_layer) # concat all embeddings merge = concatenate(em_layers) dense = Dense(10, activation='relu', kernel_initializer='he_normal')(merge) output = Dense(1, activation='sigmoid')(dense) model = Model(inputs=in_layers, outputs=output) # compile the keras model model.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy']) # plot graph plot_model(model, show_shapes=True, to_file='embeddings.png') # fit the keras model on the dataset model.fit(X_train_enc, y_train_enc, epochs=20, batch_size=16, verbose=2) # evaluate the keras model _, accuracy = model.evaluate(X_test_enc, y_test_enc, verbose=0) print('Accuracy: %.2f' % (accuracy*100)) |
Running the example prepares the data as described above, fits the model, and reports the performance.
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
In this case, the model performs reasonably well, matching what we saw for the one hot encoding in the previous section.
As the learned vectors were trained in a skilled model, it is possible to save them and use them as a general representation for these variables in other models that operate on the same data. A useful and compelling reason to explore this encoding.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 |
... Epoch 15/20 - 0s - loss: 0.4891 - acc: 0.7696 Epoch 16/20 - 0s - loss: 0.4845 - acc: 0.7749 Epoch 17/20 - 0s - loss: 0.4783 - acc: 0.7749 Epoch 18/20 - 0s - loss: 0.4763 - acc: 0.7906 Epoch 19/20 - 0s - loss: 0.4696 - acc: 0.7906 Epoch 20/20 - 0s - loss: 0.4660 - acc: 0.7958 Accuracy: 72.63 |
To confirm our understanding of the model, a plot is created and saved to the file embeddings.png in the current working directory.
The plot shows the nine inputs each mapped to a 10 element vector, meaning that the actual input to the model is a 90 element vector.
Note: Click to the image to see the large version.
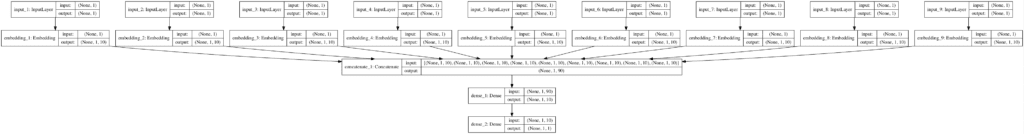
Plot of the Model Architecture With Separate Inputs and Embeddings for each Categorical Variable
Click to Enlarge.
Common Questions
This section lists some common questions and answers when encoding categorical data.
Q. What if I have a mixture of numeric and categorical data?
Or, what if I have a mixture of categorical and ordinal data?
You will need to prepare or encode each variable (column) in your dataset separately, then concatenate all of the prepared variables back together into a single array for fitting or evaluating the model.
Q. What if I have hundreds of categories?
Or, what if I concatenate many one hot encoded vectors to create a many thousand element input vector?
You can use a one hot encoding up to thousands and tens of thousands of categories. Also, having large vectors as input sounds intimidating, but the models can generally handle it.
Try an embedding; it offers the benefit of a smaller vector space (a projection) and the representation can have more meaning.
Q. What encoding technique is the best?
This is unknowable.
Test each technique (and more) on your dataset with your chosen model and discover what works best for your case.
Further Reading
This section provides more resources on the topic if you are looking to go deeper.
Posts
- Develop Your First Neural Network in Python Step-By-Step
- Why One-Hot Encode Data in Machine Learning?
- Data Preparation for Gradient Boosting with XGBoost in Python
- What Are Word Embeddings for Text?
- How to Use Word Embedding Layers for Deep Learning with Keras
- How to Use the Keras Functional API for Deep Learning
API
- sklearn.model_selection.train_test_split API.
- sklearn.preprocessing.OrdinalEncoder API.
- sklearn.preprocessing.LabelEncoder API.
- Embedding Keras API.
- Visualization Keras API.
Dataset
- Breast Cancer Data Set, UCI Machine Learning Repository.
- Breast Cancer Raw Dataset
- Breast Cancer Description
Summary
In this tutorial, you discovered how to encode categorical data when developing neural network models in Keras.
Specifically, you learned:
- The challenge of working with categorical data when using machine learning and deep learning models.
- How to integer encode and one hot encode categorical variables for modeling.
- How to learn an embedding distributed representation as part of a neural network for categorical variables.
Do you have any questions?
Ask your questions in the comments below and I will do my best to answer.






It’s midnight over here in India and My eyes are just shutting off … But the moment I read the subject of your mail… I knew I had to read to this!!
Y u ask?
For the embedding explanation . Although I’ll have my doubts lined up as and when I try it out…
… I wish to express my gratitude towards the Amazing knowledge you share with the world!
Thanks, I hope it helps on your next project!
Hi Jason,
I would like to know How to handle with multi-class and multi-label in the same dataset, for example hair color, skin color and eye color (multi-output) defined by the set of attributes for genetic markups as genes (CC, TC, etc).
If you have any kind of reference for a problem like that, I appreciate your help.
Thanks in advance.
Here is an example:
https://machinelearningmastery.com/how-to-develop-a-convolutional-neural-network-to-classify-satellite-photos-of-the-amazon-rainforest/
Hi sir,
Recently i am facing some doubt regarding how to encode a categorical data that is given in the form of bins.Actually i want to use “Age” data to build my decision tree.In some blogs it is recommended to convert continous numerical data into categorical bin data before using DT.
My doubt is after i created the bins using Discretization , how to use this bin data to build my decision Tree?? Thank in advance!
The transformed data is then used as input to the model.
Hi, Jason: Regarding “the embedding expects the categories to be ordinal encoded”, is this true for any type of entity embedding? That is, the ‘ordinal’ encoding is a must.
Excellent question!
Yes, but the mapping from labels to integers does not have to be meaningful.
I should probably have said “label encoded” or “integer encoded” to sound less scary. Sorry.
Hello,
Can you explain why this does not have to be meaningful?
I don’t know if I have understood this correctly, but what I think happens is this:
The LabelEncoder() transforms all of the words in the input column into integer values, one integer for each unique value in the input column. Which again is used as input to the embedding layer.
Won’t the embedding layer be affected by the number in the column? For example, let’s say the input words are [bad, good, great, horrible, good, good, bad, great] and the label encoder transforms this into [0, 1, 2, 3, 1, 1, 0, 2].
Does the number have no effect on the word embedding? That words that does not have resemblence is quite equal?
No, the embedding layer learns a vector representation for each word such that the “closeness” of words is meaningful/useful to the model under the chosen loss and dataset.
Although it says “expects the categories to be ordinal encoded”, the inputs are still prepared with LabelEncoder(), not OrdinalEncoder(). Why is that?
Another top question, thanks!
They both do the same thing.
Label encoder is for one column – explicitly. Ordinal encoder is for a variable number of columns.
Another question is why two Embedding objects are of different types? One is from this tutorial, and its type is “Tensor(“embedding_1/embedding_lookup/Identity_2:0″, shape=(None, 1, 5), dtype=float32)” due to functional api, and its shape is (None, 1, 5). In the other tutorial, https://machinelearningmastery.com/what-are-word-embeddings/, the Embedding object is “”, and it doesn’t even have the ‘_shape’ variable. The reason I am asking this is because in the 2nd tutorial it must use a Flatten() layer, but in 1st tutorial, it doesn’t use it. Both are embedding objects, and why their internal attributes are different?
Often embeddings are used with sequences of words as input.
Here, we have one embedding for one category. No flatten required.
Hello! Very helpful article, especially the Embedding part. One question in which I ran into during my own application:
Do you have any suggestions how to deal with a situation where the test set has unseen labels when compared to the training set? For an example below, as we fit the LabelEncoder with training data,
le.fit(X_train[:, i])
# encode
train_enc = le.transform(X_train[:, i])
—> test_enc = le.transform(X_test[:, i])
The last line would throw value error about the test set containing previously unseen labels. One option would be fitting the LabelEncoder with all data but that results into information leak which is undesirable. In reality the test set (or validation set) can certainly have previously unseen labels.
Cheers!
Thanks!
Excellent question!!!
Yes, you can remove rows with unknown labels (painful), or map unknown labels to an “unknown” vector in the embedding, typically vector at index 0 can be reserved for this – in NLP applications.
This will require more careful encoding of labels to integers, might be best to write a custom function to ensure it is consistent.
Does that help?
Hello!
Thank you for the quick and helpful response! Removing rows with unknown labels is unfortunately out of the question. I was afraid that it will require custom encoding. But it is an intriguing problem and might be crucial for my application, where I am analyzing flow-based network data and trying to encode IP-addresses and ports. An ideal solution would be, while new data points are introduced, updating the labels dynamically in a way that they can be fed to the neural net (autoencoder in this case).
Cheers, keep up the awesome work!
Let me know how you go.
Thanks.
Do you happen to have a solution for this for an XGBoost model (I am using Sklearn’s OrdinalEncoder).
A solution for what exactly?
Hi, I hope you are still responding to this thread. How exactly can you do this? Is it done when label encoding the value?
Yes, that the label encoding step to identity all unknown labels.
Hi,
The article is very helpful to understand the embedding technique. I recommend you to follow and run the examples to obtain deep perception of it.
I would like just to add that this technique is now widely used in many fields such as NLP, the biological field, image processing, especially where the data are structured as a graph by simulating the node to a word and the edge to a sentence.
Thank you Jason.
https://machinelearningmastery.com/use-word-embedding-layers-deep-learning-keras/ : the article I mentioned in my comment above !
Yes.
There are many examples for NLP, you can see some here under “word embeddings”:
https://machinelearningmastery.com/start-here/#nlp
For example:
https://machinelearningmastery.com/use-word-embedding-layers-deep-learning-keras/
hello, i’m getting an error when I try to use the last code with my dataset breast cancer. how can I fix this error? Thanks ValueError: y contains previously unseen labels: [‘clump_thickness’]
Sorry to hear that.
If you are experiencing this issue with a OneHotEncoder, you can set handle_unknown to ‘ignore’, e.g.
Hi
In this article it says:
Note: I will leave it as an exercise for you to update the example below to try specifying the order for those variables that have a natural ordering and see if it has an impact on model performance.
I tried that, and the model didn’t perform better than around 70% accuracy. Have you also tried that out? No improvement in accuracy is also what you would expect?
Another question: It’s not possible to specifying the order ONLY for those variables that have a natural ordering, you either need to specify it for all or none of them, or am I wrong? I used the categories parameter for that purpose, see:
https://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.OrdinalEncoder.html
Thanks
Very cool, thanks for trying.
No, I have not tried.
No, you can process each variable separately and union / vstack them back together prior to modeling. It’s a pain and the reason why I left it as an exercise.
how to encode a column with 10 thousand unique values stating the column to be city names ,
for unsupervised clustering thechniques
Great question!
I would recommend an learned embedding with a neural net, then compare results to other methods, like one hot, hashing, etc.
Thank you,
Any resource or solution available on Neural net embeddings?
The above tutorials shows how.
Also, I have many other examples on the blog, use the search box.
What are you having trouble with exactly?
Hi Jason,
I did not understand your suggestion
How to use “learned embedding with a neural net” to encode a column with 10 thousand unique values stating the column to be city names ?
Above example uses NN so the embedding is learned as part of training the model.
How to build an embedding for a column like city and then use it for lets say Logistic Regression
You could train the embedding in a standalone manner, e.g. via an autoencoder.
Hi Ken
I am also trying to find some thing and closes i could find is:
https://medium.com/analytics-vidhya/categorical-embedder-encoding-categorical-variables-via-neural-networks-b482afb1409d
You’re welcome.
Have you attended the NIPS conference ?
This year, no.
Not this year.
Thanks for this excellent and very useful article.
But I want to ask a question about the encoding phase. Do train and test data encoding separately is logical? For example, if a feature has different categorical values in test and train data, is it possible to trust the model? Or the model runs correctly? Maybe it will be more optimum, splitting the data as train and test after encoding…
Best regards,
You’re welcome.
The training set must be sufficiently representative of the problem – e.g. contain one example of each variable.
Hi Jason,
I am trying to apply your embedding code to some of my data. All y and x variables in the dataset(s) are string data types.
When I run the section:
in_layers = list()
em_layers = list()
for i in range(len(X_train_enc)):
# calculate the number of unique inputs
n_labels = len(unique(X_train_enc[i]))
# define input layer
in_layer = Input(shape=(1,))
# define embedding layer
em_layer = Embedding(n_labels, 10)(in_layer)
# store layers
in_layers.append(in_layer)
em_layers.append(em_layer)
I get the error:
—————————————————————————
NameError Traceback (most recent call last)
in
2 in_layers = list()
3 em_layers = list()
—-> 4 for i in range(len(X_train_enc)):
5 # calculate the number of unique inputs
6 n_labels = len(unique(X_train_enc[i]))
NameError: name ‘X_train_enc’ is not defined
Do have any ideas as to what the problem may be?
Thanks.
scott
You may have skipped some lines.
Perhaps start with the working example and slowly adapt it to use your own dataset.
Hi Jason,
I have been trying to adapt your embedding code to a multi-label classification, but have been unsuccessful. I am trying to predict 14 binary labels, using 89 categorical predictors. How would I have to change your code to account for a multi-label problem? Thank you.
Sounds great.
All of the encoding schemes are for the input variables. No change required really, you can use them directly.
Thanks for your prompt reply Jason. I am not sure exactly what you mean by “encoding schemes are for the input variables”.
Regardless, I have gone through your tutorial again, but still have not been able to figure out how to adapt it to a multi-label case.
As I mentioned, I am trying to predict 14 binary labels, with 89 features. So instead of a single outcome vector/matrix outcome nx1, I have a matrix of nx14. I am not sure how to account for this in your code.
I believe that I may be encountering problems in a few areas of your code while trying to adapt it to multi-label classification. I will try to explain these below.
I am not sure why you need to convert to 2d array in your example code below, and how I should change this if I am predicting 14, not 1, label.
– # reshape target to be a 2d array
– y = y.reshape((len(y), 1))
Also, my 14 targets are already in binary form and have a string datatype – 14 individual columns coded with 0s and 1s – so I’m not sure if I even need to include the following code. I believe I still need to generate the y_train_enc and y_test_enc, but am not sure of what the format they should be.
– # prepare target
– def prepare_targets(y_train, y_test):
– le = LabelEncoder()
– le.fit(y_train)
– y_train_enc = le.transform(y_train)
– y_test_enc = le.transform(y_test)
– return y_train_enc, y_test_enc
Also, this part confuses me. I don’t know exactly why you format the output as 3d (?array?). How would the following code be changed to account for 14 label outcomes/predictions?
– # make output 3d
– y_train_enc = y_train_enc.reshape((len(y_train_enc), 1, 1))
– y_test_enc = y_test_enc.reshape((len(y_test_enc), 1, 1))
Also, in your example you had 9 categorical features and 1 binary outcome.
I have 89 features and 14 binary outcomes. I am confused as to why you specify “10” in the code snippet below. In my case, would I have to specify 103 (89+14)?
– # define embedding layer
– em_layer = Embedding(n_labels, 10)(in_layer)
Also, what does “n_labels” actually represent
(i.e., not sure what this means – len(unique(X_train_enc[i]))),
and in the case of your example what the number would actually be?
Finally, is the “10” in your following code dictated because your example includes a total of 10 variables (9 features and 1 outcome)?
– dense = Dense(10, activation=’relu’, kernel_initializer=’he_normal’)(merge)
In the end, I also want to be able to produce an nx14 pd dataframe that contains the class (0/1, not probabilities) predictions for all 14 labels, from which I can use scikit learn to produce multi-label performance metrics.
I have not been able to find another example that comes close to doing what I need to do – namely use categorical feature embedding in a multi-label classification case.
I greatly appreciate you taking the time to answer my questions. I have always found your tutorials and posts extremely valuable.
Scott
That is a huge comment, I cannot read/process it all.
It sounds like you are trying to encode the target variable rather than the the input variable. This tutorial is focused on encoding the input variables.
If you want to encode a target variable with n classes for multi-label classification, you must use this:
https://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.MultiLabelBinarizer.html
Jason,
Sorry for the long comment.
I am not trying to encode the target – I have 14 binary target columns – 14 labels.
Perhaps you can answer just this one question.
Why do you format the output as 3d (?array?). How would the following code from your example be changed to handle 14 binary label outcomes/predictions (as opposed to your example which is classification of 1 binary label?
– # make output 3d
– y_train_enc = y_train_enc.reshape((len(y_train_enc), 1, 1))
– y_test_enc = y_test_enc.reshape((len(y_test_enc), 1, 1))
My guess is that I would change the last 1 to 14:
– y_train_enc = y_train_enc.reshape((len(y_train_enc), 1, 14))
– y_test_enc = y_test_enc.reshape((len(y_test_enc), 1, 14))
Am I correct?
Thanks again.
Scott
We don’t make any output arrays 3d in this post. Are you referring to a different post perhaps?
For some models, like encoder-decoder models we need to have a 3d output, e.g. an output sequence for each input sequence. I think this is what you are referring to.
If so, you would have n samples, t time steps, and f features, where f features would be the 14 labels.
Jason.
Then what do lines 59-61 in your full embedding code do?
They seem to indicate converting 2d to 3d. Below is the code lines 59-61 in your embedding example.
# make output 3d
y_train_enc = y_train_enc.reshape((len(y_train_enc), 1, 1))
y_test_enc = y_test_enc.reshape((len(y_test_enc), 1, 1))
Thanks again.
Ah I see. Thanks, I missed that.
Umm, I think the embedding should be flattened before going into the dense. We don’t so the structure stays 3d all the way to output. It’s not really 3d, just 1 time step and 1 feature per sample.
You could try and wrestle with the 3d output or try adding a flatten layer after the concat embeddings. I think that would do try trick off the top of my head. I believe I did that when working with nlp models:
Jason,
I figured my main problem it out. I just needed to change the 3rd dim from 1 to 14 here, assuming I have 14 labels to predict.
Just a FYI – below is part of the code I used to run the embedding model and then calculate multilabel metrics (using sklearn.metrics).
Starting with 14 labels and 89 predictors, as pandas dataframes:
Hope example is of use to somebody
Thanks again for you help Jason.
scott
Well done!
Thanks for sharing.
Hi, again, Jason,
I appears that you use 10 as the embedding dimension, in your “prepare head” section of code. Does it mean that you are using 10 as the dimension for ALL the categorical variables?
If so, I would like to assign a different embedding dimension for each categorical variable, given the number of levels in each categorical variable can be different. I think my following code would do this but I’m not sure it integrates with your code properly – note the key part I added is “cat_embsizes[cat]” which replaces your entry of “10”, which represents the embedding dimension. Here I calculate the number of unique values of each categorical variable, calculate dimension I want to use for each variable, then apply that list to your “prepare head” code. Please let me know what you think – will this work? Thanks again.
Yes.
Good idea!
Sorry, I cannot debug/review this for you:
https://machinelearningmastery.com/faq/single-faq/can-you-read-review-or-debug-my-code
I think the sentence “In this case, we will ignore any possible existing ordinal relationship and assume all variables are categorical. It can still be helpful to use an ordinal encoding, at least as a point of reference with other encoding schemes.” is out of its place.
Am I wrong?
How so?
hi. i want to ask you how to split x and y variable without ‘,’ which i take the data from excel without delimeter. i always got an error on this type
ValueError: Error when checking target: expected dense_18 to have shape (1,) but got array with shape (31,)
need an explanation on this. thank you
Perhaps this will help you load your data:
https://machinelearningmastery.com/load-machine-learning-data-python/
Thanks for the info Jason 🙂
Hi Jason,
Awesome post thanks!
I want to ask something, I was trying to make the hot and ordinal encode and , but my dataseth has both type variables, categoricals and continuos, so as you said , for the ordinal I preprocessed each column and concatenate all the variables back together into a single array, and it works, but doing the same with hot encode , didn’t work, when I try to make “np.concatenate((x_cont,x_cat)” I got an error “”all the input arrays must have same number of dimensions, but the array at index 0 has 2 dimension(s) and the array at index 1 has 0 dimension(s)”” and I know how to solve.
Thanks!
Thanks!
Good question, you can use the “ColumnTransformer” to handle encoding two different variables types, here is an example:
https://machinelearningmastery.com/columntransformer-for-numerical-and-categorical-data/
Hi Jason,
Other question concerning cases with numeric and categorical data with the Learned Embedding technique: do I need to add an input layer to the model for the numerical data (in addition to the layers for the categorical variables)?
Thanks!
Yes. A separate input for other data is a great idea, e.g. a multi-input model:
https://machinelearningmastery.com/keras-functional-api-deep-learning/
Hello Jason,
Excellent post and much appreciated.
I noticed that when I was reviewing the sklearn documentation regarding LabelEncoer
(https://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.LabelEncoder.html)
the docs say:
“This transformer should be used to encode target values, i.e. y, and not the input X.”
In this example, you haven’t hesitated in using the LabelEncoder on inputs. Do you know what the problem is with using LabelEncoder for inputs or why sklearn cautions against this? (I am trying to figure out what limitations it may cause to my model.)
Thank you!
Thanks.
Yes, you’re supposed to use the ordinal encoder for input instead. They do the same thing. I used it here because we are working with 1d data, like a target.
For the sake of an example, suppose we have 3 target categories: up, down, or stay. As an alternative, can we programmatically map these target categories to 0, 1, or 2 as opposed to running OneHotEncoder()/OrdinalEncoder() on it? Thanks for your work!
Yes.
Thank you!
I’m trying to make it work for a regression model but I get the following error (after one epoch) which happens to shift from run to run. I have tried to increase the input shape for the embedding by the number of features +1 etc. but could not make it work.
Thank you for your help!
Error message:
InvalidArgumentError: indices[3,0] = 5 is not in [0, 5)
[[node model_51/embedding_243/embedding_lookup (defined at :99) ]] [Op:__inference_distributed_function_31433]
Errors may have originated from an input operation.
Input Source operations connected to node model_51/embedding_243/embedding_lookup:
model_51/embedding_243/embedding_lookup/29082
On the next run that message would change to:
InvalidArgumentError: indices[0,0] = 1 is not in [0, 1)
[[node model_52/embedding_325/embedding_lookup (defined at :99) ]] [Op:__inference_distributed_function_38501]
Errors may have originated from an input operation.
Input Source operations connected to node model_52/embedding_325/embedding_lookup:
model_52/embedding_325/embedding_lookup/36354
Embedding expect integer encoding inputs only. Not floating point values.
Thank you! It turned out to be another issue: I had to set the ‘n_labels’ to the length of category with the highest number of unique labels +1 (in my case 16) for all embeddings. I hope this is not altering the model.
Interesting.
Hello Sir,
Thanks this content is great! I have a problem where I have categorical variables grouped in bins: ‘0 years’ , ‘1-3 years’ , ‘3-5 years’ , ‘5 or more years’. I think I should not use the ordinal encoding because then it will seem like they are different by one step (1,2,3) but the one hot encoding might miss the natural ordering of this bins.
What do you recommend ?
I recommend exploring both and use the approach that results in the best performance for your chosen model and test harness.
Ordinal encoding with specified order sounds appropriate.
Thankyou very much for your response. In the example mentioned what do you recommend to use for ordinal encoding?
0 years : 0
1-3 years : 1
3-5 years: 2
And such would be a correct approach?
The only “correct” approach is the one you try that performs better than others.
I recommend prototyping a number of different approaches and discover what works best for your specific data and choice of model.
Ok thankyou.
You’re welcome.
Hi,
I have a question…my data has a mix of categorical and continuous variables. The target variable is a multi class variable (and not binary). It has to predict 3 different classes. I have one hot encoded categorical variables. Should I encode target variable also, bcoz if I do there will be 3 separate columns (with values 0-1) and I am not sure how the prediction will look like then??
Yes, the target should be encoded.
Hi Jason,
Thank you very much for this info.
I wonder if you know why running
merge = concatenate(em_layers)
resulted with the following error:
ValueError: zero-dimensional arrays cannot be concatenated
Sorry to hear that you are having an error, I have some suggestions here that may help:
https://machinelearningmastery.com/faq/single-faq/why-does-the-code-in-the-tutorial-not-work-for-me
Hi Jason!
I have read a lot about your features encoding posts, they have been a great help!
but I have a big doubt. What is the best method to encode categorical features with a big amount of categories, i.e., more than 500 categories in only one feature? I have tried LabelBinarizer from sklearn, but I’m not sure is I’m doing right!
Thanks a lot in advance.
There is no objective best method, you must discover what works best for your dataset, here are some ideas to try:
https://machinelearningmastery.com/faq/single-faq/how-do-i-handle-a-large-number-of-categories
Hi Jason, if I have categorical attributes in my dataset that consists of multiple categorical values (say, around 1000. For eg. a column ‘Name’ there are 1000 different names), label encoding would create 1000 integer values.
So, now, if i want to train a neural net on the dataset, should i use this column in this raw (label encoded) format, or is there any way I can reduce the values? Is it okay to feed into the NN such large values, especially when some other attribute may be having values in the range (0-10)?
One Hot encoding would make the feature vector very sparse. So, don’t know if it would be correct to do that
You could scale the values, e.g. normalize.
Also compare results to a one hot encoding and an embedding. I would expect an embedding to perform well.
A one hot encoding would be sparse, perhaps try it any way to confirm how it performs.
Hi Jason:
Great Tutorial !. I esteem in deep, all of your teachings. Thanks!
If you allow me, I will share some comments and questions:
COMMENTS:
1) I experimented applying “coding” conversions to the whole (X, Y) categorical dataset, before splitting (X,Y) “tensors” into train and test groups. So, making sure I gather all possible dataset labels or categorical types are collected, but, on the other hand applying “embedding” coding, before splitting require to have to convert a list of array into numpy and transpose it, and viceversa before to feed the input embedding model.
By the way, when applying “embedding” coding I could not apply “stratify” option, to the “train_test_split()” function, the code crash it!. I do not know why.
2) I experimented applying new “tf.keras” API, but when I define “Concatenate” layer, it seems to work differently from “keras.layers.merge. concatenate”, and also the code crash. So I return to import all libraries from keras standalone, such your “keras.layers.merge.concatenate”.
3) I experimented adding “batchnormalization” and “dropout” layers, as part of the fully connected model head. I apply also “Kernel_regularizer “on dense layers.
But without any significant accuracy improvement!.
4) I experimented adding “Conv1D” (kernel_size=1) and “MaxPool1D”(pool_size=1) after embedding layer definition, and also applying Flatten layer conversion before concatenate layer.
But not significant change was founded!.
5) I apply KFold() function form Sklearn library, beside to repeat same training in order to perform statistically impact on different train-test validation.
6) Taking into account that dataset is clearly Imbalanced (81 recurrence vs. 205 non-recurrence labels), I am surprised that applying class-weight () argument to train model (.fit() method), not only the accuracy did not improve but even it gets worse, from 72% to 68.% accuracy .
I do not know why. !
7) I got the best encoding results applying “OneHotEncoder” (76%), followed by “OrdinalEncoder” (75%), and finally by Embedding (72%). In spite OneHotEncoder and OrdinalEncoder are much more simple techniques than applying Embedding coding!
8) I share same comments of some of the responses said i this timeline , in the sense that explain better the dimensions change of input tensor X, Y, it is hard to follow (in the tutorial) beside some list to numpy conversions including on embedding layers implementation.
As far as I know, the summary of encoding could be:
when applying “OrdinalEncoder” we convert categorical labels to integers and, the 9 original features of dataset it is invariant as model input [286, 9]. But, when applying “OneHotencoder”, that convert categorical into many 0’s and an unique 1 then, the input feature it is expanded from 9 to 43, so the input model dimensions are now [286, 43].
And finally when applying “Embedding encoding and deep learning layers” it is retained 2D original dimensions [286, 9] on input features, but in the embedding layer it is introduced an extra dimension (associated to the new embedding vector dimension e.g. 10), so now de total dimensions becomes 3D [286 , 9, 10]
QUESTIONS:
a) Why on NLP techniques (e.g. sentiment analysis text classification) we can encode the words (that are the different features of the text) on a single embedding layer (common to all input) but now, we have to introduce a single input + embedding layer per each feature?
Is it possible to apply here, only an embedding layer for all the input features, such as on NLP text classification ?
b) On NLP techniques we have to use always flatten layer to present all feature extraction to the fully connected head model, from this deep layers, why it is not (obligatory) to apply here ?
c) When applying dimensions reductions methods such as PCA, feature selection, etc. it is clear that we want to focus on making the input more simple and learning more efficient…but when we apply embedding, Onehot, or other deep learning layers (convolutional, etc.) it seems to add new extra dimensions (vectors, tensors) where to project the new feature extractions…so conceptually seems two ideas (dimensions expansion and feature reduction) working on opposite directions :-(!!
sorry for too extensive text !
regards
JG
Stratify needs a label to stratify by, one column with a limited number of values.
Wow, lots of cool tests! You could also compare different embedding sizes.
In NLP, we only have one “variable”. Here we have many different variables.
Depends on the model as to whether a flatten layer is required. E.g. a vector output from embedding can be used directly by a dense layer.
I see PCA and embedding doing the same kind of thing. Projecting to a new vector space that preserves relationships between observations.
Hi, I am working on a similar problem and trying to stratify my y_train. I have onehotencoded my data. Do I have to change the “stratify = y_train” in
“X_train, X_val, y_train, y_val = train_test_split(X_train, y_train, test_size = 0.15, shuffle = True, stratify = y_train)”
because it makes no real difference when I do add it.
You must split the data first, then encode it.
thks
Hi Jason,
I’m a green hand of deep learning, and I have a data set whose 60 variables are all binary variables (0 for “good” and 1 for “bad”). In this case, do I need to use the ways you shared in this post to transfer my binay variables further? Or shall I just use the original dataset for CNN-based classification?
Thanks a lot!
Probably not.
If it is a tabular dataset, then a CNN would not be appropriate, instead you would use an MLP, more here:
https://machinelearningmastery.com/when-to-use-mlp-cnn-and-rnn-neural-networks/
Got it, it really helps, appreciate it!
You’re welcome.
Hi Jason – I came across a methodology to convert categorical data into normal distribution using following procedure: are you aware of how this would be implemented in python?
The discrete columns are encoded into numerical [0,1] columns using the following method.
1. Discrete values are first sorted in descending order based on their proportion in the dataset.
2. Then, the [0,1] interval is split into sections [ac,bc] based on the proportion of each category c.
3. To convert a discrete value to a numerical one, we replace it with a value sampled from a Gaussian distribution centred at the midpoint of [ac,bc] and with standard deviation σ = (bc − ac)/6.
I’ve not seen this, sorry.
if i encoding the variable blood group having four levels A,B,AB and O .to perform encoding i wish to drop two levels AB,O. suggest suitable encoding which will represent the four levels
Perhaps test different encoding schemes and discover what works best for your chosen model.
Try this:
A B
A 1 0
B 0 1
AB 1 1
O 0 0
“You will need to prepare or encode each variable (column) in your dataset separately, then concatenate all of the prepared variables back together into a single array for fitting or evaluating the model.”
I recently read https://www.cs.otago.ac.nz/staffpriv/mccane/publications/distance_categorical.pdf, which is a great paper about simple distance functions for mixed variables like this. One of their suggestions is to encode unrelated variables as regular simplex coordinates. Technically it’s possible to do this, as they suggest, in one fewer dimensions than there are choices for your categorical variable. Actually one-hot encoding is really just simplex vertices using the same number of dimensions as there are choices: http://www.math.brown.edu/~banchoff/Beyond3d/chapter8/section03.html.
But doesn’t finding a whole vector for some input variable, concatenating it with continuous variables that don’t take up so many inputs, and sending through a model lead to some kind of imbalance? Seems like the model would naturally put more weight on the thing that’s taking up all this input space, and it would have to learn to consider those single-entry numerical inputs more important.
Thanks for sharing.
Depends on the choice of model.
Hello Jason,
Thank you for another great topic on dealing with categorical data and embeddings.
One question,
loop …
em_layer = Embedding(n_labels, 10)(in_layer)
merge = concatenate(em_layers)
I can see in the above code you have chosen a constant embedding vector size of 10 for all categorical features. But in reality, this may not be the case, one can have different size vectors depending upon the number of unique categories of each feature.
For such a case directly concatenating all the embedding layers fail, so,
em_layer = Embedding(n_labels, 10)(in_layer)
em_layer = Reshape((10, ))(em_layer)
Does the above code makes sense, or can you propose any different solution?
You’re welcome.
Yes, you would create one embedding layer per input variable. Each layer has the same input layer.
You do not want to stack embedding layers have you have done in your code.
Sorry I did not get the last sentence.
What do you mean by do not want to stack?
Here is my full code, the model compile well, but have the embedding layers logically structured correctly?
def emb_sz_rule(n_cat:int)->int: return min(600, round(1.6 * n_cat**0.56))
embedding_inputs=[]
embedding_output=[]
non_embedding_inputs = [Input(shape=(continuous_list.__len__(),))]
for name in categorical_list:
nb_unique_classes = df_dataframe[name].unique().size
embedding_size = emb_sz_rule(nb_unique_classes)
# One Embedding Layer for each categorical variable
model_inputs = Input(shape=(1,))
model_outputs = Embedding(nb_unique_classes, embedding_size)(model_inputs)
model_outputs = Reshape(target_shape=(embedding_size,))(model_outputs)
embedding_inputs.append(model_inputs)
embedding_output.append(model_outputs)
model_layer = concatenate(embedding_output + non_embedding_inputs)
model_layer = Dense(128, activation=’relu’)(model_layer)
model_layer = BatchNormalization()(model_layer)
model_layer = Dropout(0.5)(model_layer)
model_layer = Dense(64, activation=’relu’)(model_layer)
model_layer = BatchNormalization()(model_layer)
model_layer = Dropout(0.5)(model_layer)
model_outputs = Dense(1, activation=’sigmoid’)(model_layer)
model = Model(inputs=embedding_inputs + non_embedding_inputs, outputs=model_outputs)
optim = Adam()
model.compile(loss=’binary_crossentropy’, optimizer=optim, metrics=[‘accuracy’])
Never-mind, I must have misread your code – I thought you were linking/stacking one embedding to another – which would be madness.
# prepare target
def prepare_targets(y_train, y_test):
# print(“y_train -“, y_train)
# print(“y_test -“, y_test)
le = LabelEncoder()
le.fit(y_train)
y_train_enc = le.transform(y_train)
y_test_enc = le.transform(y_test) <–error
return y_train_enc, y_test_enc
Error – ValueError: y contains previously unseen labels: 358
My csv data – text1, text2, euclidean dist
Perhaps ensure that your training dataset is a representative sample of your problem.
After reading the tutorial and some of the comments it is not clear for me how to proceed in case of a mix categorical and numerical features:
Procedure 1:
1.1 split datase in categorical and numerical
1.2 do PCA over the numerical
1.3 do hot encoding or embending over the categorical
1.4 concact both datasets
Procedure 2:
2.1 split datase in categorical and numerical
2.2 do hot encoding or embending over the categorical
2.3 concact both datasets
2.4 do PCA over all datasets
how would you do?
If you are using sklearn models, you can use a columntransformer object to handle each data type with separate pipelines.
If you are using keras models, you can prepare each variable or group of variables separately and either concat the prepared columns or have a separate input model for each variable or group of variables.
Hi, How to get the original labels of predicted values after encoding ?
If you used an ordinal encoder to encoder strings to integers, use the same object with inverse_transform() to convert integers back to strings.
https://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.OrdinalEncoder.html#sklearn.preprocessing.OrdinalEncoder.inverse_transform
Thanks for the great article, Jason.
How can I concatenate a prepared variable (one-hot encoding, i.e. in Compressed Sparse Row format) with a purely numerical variable that required no preparation)?
–Dean
You’re welcome.
Perhaps numpy concatenate() or hstack(), see examples here:
https://machinelearningmastery.com/gentle-introduction-n-dimensional-arrays-python-numpy/
Thanks!
Actually, I just concatenated the vector for the prepared ohe set (sparse = False) with the pure numerical vectors and it worked just fine.
I’m happy to hear you’re making progress!
Hi Jason
When using embedding, if during test, it encounter never seen data, the
mapping(“coordinate”) will not be updated, But this will lead very big error right?
supposed super simple example , in the beginning we have 2 data train, and 2 dimension embed:
1 —> [0.21, -0.3] (value 1 –> converted to 2 dimension of embedding)
2 —> [0.51, -0.5] (value 2 –> converted to 2 dimension of embedding)
After training several epoch, value of embedding is learned/thus changed
1 —> [0.25, -0.31] (value 1 –> converted to 2 dimension of embedding)
2 —> [0.26, -0.32] (value 2 –> converted to 2 dimension of embedding)
So after learn, embedding position 2 vector very close each other
So new test data is coming, value : 3 —> [ 0.5, 0.7] , this mapping is
since never learned, so the vector of mapping is very different(far) apart
with the training data, where intuitively it should close each other
So is it natural? or any thing that we can do?
If you are using embeddings and the model receives a token not seen during training, it is mapped to input 0 “unknown”.
It is not a disaster, quite common in NLP problems.
You must ensure your training data is reasonably representative of the problem.
—————————————————————————
ValueError Traceback (most recent call last)
in ()
46 X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=1)
47 # prepare input data
—> 48 X_train_enc, X_test_enc = prepare_inputs(X_train, X_test)
49 # prepare output data
50 y_train_enc, y_test_enc = prepare_targets(y_train, y_test)
in prepare_inputs(X_train, X_test)
28 def prepare_inputs(X_train, X_test):
29 ohe = OneHotEncoder(sparse=False, handle_unknown=’ignore’)
—> 30 ohe.fit(X_train)
31 X_train_enc = ohe.transform(X_train)
32 X_test_enc = ohe.transform(X_test)
~\Anaconda3\lib\site-packages\sklearn\preprocessing\data.py in fit(self, X, y)
1954 self
1955 “””
-> 1956 self.fit_transform(X)
1957 return self
1958
~\Anaconda3\lib\site-packages\sklearn\preprocessing\data.py in fit_transform(self, X, y)
2017 “””
2018 return _transform_selected(X, self._fit_transform,
-> 2019 self.categorical_features, copy=True)
2020
2021 def _transform(self, X):
~\Anaconda3\lib\site-packages\sklearn\preprocessing\data.py in _transform_selected(X, transform, selected, copy)
1807 X : array or sparse matrix, shape=(n_samples, n_features_new)
1808 “””
-> 1809 X = check_array(X, accept_sparse=’csc’, copy=copy, dtype=FLOAT_DTYPES)
1810
1811 if isinstance(selected, six.string_types) and selected == “all”:
~\Anaconda3\lib\site-packages\sklearn\utils\validation.py in check_array(array, accept_sparse, dtype, order, copy, force_all_finite, ensure_2d, allow_nd, ensure_min_samples, ensure_min_features, warn_on_dtype, estimator)
431 force_all_finite)
432 else:
–> 433 array = np.array(array, dtype=dtype, order=order, copy=copy)
434
435 if ensure_2d:
ValueError: could not convert string to float: ‘Heat (1995)’
###############################
how to fix above error?
Perhaps these tips will help:
https://machinelearningmastery.com/faq/single-faq/why-does-the-code-in-the-tutorial-not-work-for-me
# example of learned embedding encoding for a neural network
from numpy import unique
from pandas import read_csv
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import LabelEncoder
from keras.models import Model
from keras.layers import Input
from keras.layers import Dense
from keras.layers import Embedding
from keras.layers.merge import concatenate
from keras.utils import plot_model
# load the dataset
filename=’mer.csv’
def load_dataset(filename):
# load the dataset as a pandas DataFrame
data = read_csv(filename, header=None)
data=data.drop([0])
data=data.dropna()
# retrieve numpy array
dataset = data.values
# split into input (X) and output (y) variables
X = dataset[:, :-1]
y = dataset[:,-1]
# format all fields as string
X = X.astype(str)
# reshape target to be a 2d array
y = y.reshape((len(y), 1))
return X, y
# prepare input data
def prepare_inputs(X_train, X_test):
X_train_enc, X_test_enc = list(), list()
# label encode each column
for i in range(X_train.shape[1]):
le = LabelEncoder()
le.fit(X_train[:, i])
# encode
train_enc = le.transform(X_train[:, i])
test_enc = le.transform(X_test[:, i])
# store
X_train_enc.append(train_enc)
X_test_enc.append(test_enc)
return X_train_enc, X_test_enc
# prepare target
#from sklearn.preprocessing import MultiLabelBinarizer
def prepare_targets(y_train, y_test):
#le=MultiLabelBinarizer()
le = LabelEncoder()
le.fit(y_train)
y_train_enc = le.transform(y_train)
y_test_enc = le.transform(y_test)
return y_train_enc, y_test_enc
# load the dataset
X, y = load_dataset(r’C:\Users\sulai\PycharmProjects\project88\mer.csv’)
# split into train and test sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=1)
# prepare input data
X_train_enc, X_test_enc = prepare_inputs(X_train, X_test)
# prepare output data
y_train_enc, y_test_enc = prepare_targets(y_train, y_test)
# make output 3d
y_train_enc = y_train_enc.reshape((len(y_train_enc), 1, 1))
y_test_enc = y_test_enc.reshape((len(y_test_enc), 1, 1))
# prepare each input head
in_layers = list()
em_layers = list()
for i in range(len(X_train_enc)):
# calculate the number of unique inputs
n_labels = len(unique(X_train_enc[i]))
# define input layer
in_layer = Input(shape=(1,))
# define embedding layer
em_layer = Embedding(n_labels, 10)(in_layer)
# store layers
in_layers.append(in_layer)
em_layers.append(em_layer)
# concat all embeddings
merge = concatenate(em_layers)
dense = Dense(10, activation=’relu’, kernel_initializer=’he_normal’)(merge)
output = Dense(10, activation=’softmax’)(dense)
model = Model(inputs=in_layers, outputs=output)
# compile the keras model
model.compile(loss=’categorical_crossentropy’, optimizer=’adam’, metrics=[‘accuracy’])
# plot graph
plot_model(model, show_shapes=True, to_file=’embeddings.png’)
# fit the keras model on the dataset
model.fit(X_train_enc, y_train_enc, epochs=20, batch_size=16, verbose=2)
# evaluate the keras model
_, accuracy = model.evaluate(X_test_enc, y_test_enc, verbose=0)
print(‘Accuracy: %.2f’ % (accuracy*100))
this is your code. just I changed dataset
#############################
Thanks for sharing.
ValueError Traceback (most recent call last)
in ()
59 X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=1)
60 # prepare input data
—> 61 X_train_enc, X_test_enc = prepare_inputs(X_train, X_test)
62 # prepare output data
63 y_train_enc, y_test_enc = prepare_targets(y_train, y_test)
in prepare_inputs(X_train, X_test)
38 # encode
39 train_enc = le.transform(X_train[:, i])
—> 40 test_enc = le.transform(X_test[:, i])
41 # store
42 X_train_enc.append(train_enc)
~\Anaconda3\lib\site-packages\sklearn\preprocessing\label.py in transform(self, y)
131 if len(np.intersect1d(classes, self.classes_)) 133 raise ValueError(“y contains new labels: %s” % str(diff))
134 return np.searchsorted(self.classes_, y)
135
ValueError: y contains new labels: [‘120’ ‘137’ ‘145’ ‘147’ ’15’ ‘159’ ‘174’ ‘195’ ‘208’ ‘214’ ‘222’ ’23’
’24’ ‘248’ ‘259’ ‘264’ ‘289’ ‘290’ ‘291’ ‘298’ ‘301’ ‘328’ ‘331’ ’34’
‘341’ ‘354’ ‘361’ ‘367’ ‘378’ ‘401’ ‘402’ ‘410’ ‘441’ ‘446’ ’46’ ‘468’
‘485’ ‘489’ ‘5’ ‘510’ ‘512’ ‘513’ ‘520’ ‘533’ ’54’ ‘550’ ‘579’ ‘586’
‘587’ ’59’ ’73’ ’79’ ’89’ ’90’ ’99’]
#################
Hi Jason,
sorry, I mentioned above code. How to fix above errors.
Perhaps these tips will help:
https://machinelearningmastery.com/faq/single-faq/can-you-read-review-or-debug-my-code
# example of learned embedding encoding for a neural network
from numpy import unique
from pandas import read_csv
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import LabelEncoder
from sklearn.preprocessing import MultiLabelBinarizer
from keras.models import Model
from keras.layers import Input
from keras.layers import Dense
from keras.layers import Embedding
from keras.layers.merge import concatenate
from keras.utils import plot_model
# load the dataset
def load_dataset(filename):
# load the dataset as a pandas DataFrame
data = read_csv(filename, header=None)
data = data.drop([0])
data = data.dropna()
# retrieve numpy array
dataset = data.values
# split into input (X) and output (y) variables
X = dataset[:, :-1]
y = dataset[:,-1]
# format all fields as string
X = X.astype(str)
# reshape target to be a 2d array
y = y.reshape((len(y), 1))
return X, y
# prepare input data
def prepare_inputs(X_train, X_test):
X_train_enc, X_test_enc = list(), list()
# label encode each column
for i in range(X_train.shape[1]):
le = LabelEncoder()
le.fit(X_train[:, i])
# encode
train_enc = le.transform(X_train[:, i])
test_enc = le.transform(X_test[:, i])
# store
X_train_enc.append(train_enc)
X_test_enc.append(test_enc)
return X_train_enc, X_test_enc
# prepare target
def prepare_targets(y_train, y_test):
le = MultiLabelBinarizer()
le.fit(y_train)
y_train_enc = le.transform(y_train)
y_test_enc = le.transform(y_test)
return y_train_enc, y_test_enc
# load the dataset
X, y = load_dataset(r’C:\Users\sulai\PycharmProjects\project88\mer.csv’)
# split into train and test sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=1)
# prepare input data
X_train_enc, X_test_enc = prepare_inputs(X_train, X_test)
# prepare output data
y_train_enc, y_test_enc = prepare_targets(y_train, y_test)
# make output 3d
y_train_enc = y_train_enc.reshape((len(y_train_enc), 1, 10))
y_test_enc = y_test_enc.reshape((len(y_test_enc), 1, 10))
# prepare each input head
in_layers = list()
em_layers = list()
for i in range(len(X_train_enc)):
# calculate the number of unique inputs
n_labels = len(unique(X_train_enc[i]))
# define input layer
in_layer = Input(shape=(1,))
# define embedding layer
em_layer = Embedding(n_labels, 10)(in_layer)
# store layers
in_layers.append(in_layer)
em_layers.append(em_layer)
# concat all embeddings
merge = concatenate(em_layers)
dense = Dense(10, activation=’relu’, kernel_initializer=’he_normal’)(merge)
output = Dense(10, activation=’softmax’)(dense)
model = Model(inputs=in_layers, outputs=output)
# compile the keras model
model.compile(loss=’categorical_crossentropy’, optimizer=’adam’, metrics=[‘accuracy’])
# plot graph
plot_model(model, show_shapes=True, to_file=’embeddings.png’)
# fit the keras model on the dataset
model.fit(X_train_enc, y_train_enc, epochs=20, batch_size=16, verbose=2)
# evaluate the keras model
_, accuracy = model.evaluate(X_test_enc, y_test_enc, verbose=0)
print(‘Accuracy: %.2f’ % (accuracy*100))
##############################################
ValueError Traceback (most recent call last)
in ()
57 X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=1)
58 # prepare input data
—> 59 X_train_enc, X_test_enc = prepare_inputs(X_train, X_test)
60 # prepare output data
61 y_train_enc, y_test_enc = prepare_targets(y_train, y_test)
in prepare_inputs(X_train, X_test)
38 # encode
39 train_enc = le.transform(X_train[:, i])
—> 40 test_enc = le.transform(X_test[:, i])
41 # store
42 X_train_enc.append(train_enc)
~\Anaconda3\lib\site-packages\sklearn\preprocessing\label.py in transform(self, y)
131 if len(np.intersect1d(classes, self.classes_)) 133 raise ValueError(“y contains new labels: %s” % str(diff))
134 return np.searchsorted(self.classes_, y)
135
ValueError: y contains new labels: [‘102’ ‘120’ ‘137’ ‘145’ ‘147’ ’15’ ‘159’ ‘174’ ‘195’ ‘208’ ‘214’ ‘222’
‘223’ ’23’ ’24’ ‘247’ ‘248’ ‘259’ ‘264’ ‘286’ ‘289’ ‘290’ ‘291’ ‘298’
‘301’ ‘325’ ‘328’ ‘331’ ’34’ ‘341’ ‘354’ ‘361’ ‘367’ ‘378’ ‘401’ ‘402’
‘410’ ‘426’ ‘441’ ‘446’ ‘452’ ’46’ ‘468’ ‘485’ ‘489’ ‘5’ ‘510’ ‘512’
‘513’ ‘520’ ‘533’ ’54’ ‘550’ ‘579’ ‘586’ ‘587’ ’59’ ‘603’ ‘605’ ’73’ ’79’
’89’ ’90’ ’99’]
#############
please help me, I have multiclass problem i.e 10 classes.dataframe involed in categorical column and numerical columns. there are some observations in test data.it is not existing in training process.
Perhaps you can configure your data preparation step to ignore new categorical values not seen in the training dataset.
I know that the OneHotEncoder class can be configured do this, perhaps the other methods can too?
Under “Learned Embedding”, you mention “…prepare the input data using an ordinal encoding…” but you use LabelEncoder in your code. Any reason why you didn’t use OrdinalEncoder? Thanks.
They are the same thing.
Thanks for the article!
When using an embedding layer as an input feature in another model (say XGBoost), how can it be concatenated with continuous features when the dimensions differ from the original input data? An embedding layer has the dimensions (nr. unique categories, chosen output size).
You’re welcome!
You can prepare the data up front using the embedding and use numpy functions to concat the vectors together.
I mean that the extracted embeddings have different dimensions to the original input data, so how do we join these differently sized arrays/ dataframes to create the input to a seperate model? If each row in the embedding array represents a category, I assume that we can inner join the embeddings array to our training samples on category, but how can we be certain which category is represented by a given row in the embedding array?
Do you maybe know of a code example where embeddings are used as inputs to a seperate model of a different type?
An update:
I find that we must specify:
em_layer = Embedding(n_labels + 1, output_size)(in_layer)
to prevent an out-of-range error such as:
tensorflow.python.framework.errors_impl.InvalidArgumentError: indices[157,0] = 52 is not in [0, 52)
This results in an embedding of size (n_labels + 1, output_size): there is one more row in the embedding than there are categories/ labels. This adds to the puzzle of how to map a row in an embedding back to a category.
Please help!
Perhaps ensure you are using Python 3.6, Keras 2.4 and TensorFlow 2.4.
The embedding will be a vector, one vector per input sample. This vector can be concatenated with other input variables for the sample to create a new input sample.
There are many numpy functions for concatenating vectors.
Got it! My mistake was extracting the embedding layer weights rather than the embedding layer outputs.
To get the outputs with shape (nr. samples, chosen output size), name the embedding layer as follows:
em_layer = Embedding(n_labels, 10, name=f”embedding_{i}”)(in_layer)
and finally, retreive the outputs with:
intermediate_layer_model = Model(inputs=model.input,
outputs=model.get_layer(’embedding_1′).output)
embedding_1_output = intermediate_layer_model.predict(X_train_enc)
print(embedding_1_output.shape)
# (191, 1, 10)
intermediate_output = \
embedding_1_output.reshape(embedding_1_output.shape[0], -1)
print(intermediate_output.shape)
# (191, 10)
Well done!
I got the following warning for One Hot Encode. What should I do?
C:\Users\Jalal\Anaconda3\lib\site-packages\tensorflow\python\framework\indexed_slices.py:449: UserWarning: Converting sparse IndexedSlices(IndexedSlices(indices=Tensor(“gradient_tape/sequential_1/dense_2/embedding_lookup_sparse/Reshape_1:0”, shape=(None,), dtype=int32), values=Tensor(“gradient_tape/sequential_1/dense_2/embedding_lookup_sparse/Reshape:0”, shape=(None, 10), dtype=float32), dense_shape=Tensor(“gradient_tape/sequential_1/dense_2/embedding_lookup_sparse/Cast:0”, shape=(2,), dtype=int32))) to a dense Tensor of unknown shape. This may consume a large amount of memory.
“shape. This may consume a large amount of memory.” % value)
Just a warning, not error. It may not really impact you. You can simply ignore it, or as it said, if you run out of memory, then you need to do something to work around that.
\lib\site-packages\sklearn\utils\validation.py:63: DataConversionWarning: A column-vector y was passed when a 1d array was expected. Please change the shape of y to (n_samples, ), for example using ravel().
return f(*args, **kwargs)
This error is just as it said, pass in y.ravel() instead of y
Hi i’m using your code for my own data and i got the following error, what should i do?
X_train_enc, X_test_enc = prepare_inputs(X_train, X_test)
# prepare output data
—-> y_train_enc, y_test_enc = prepare_targets(y_train, y_test)
in prepare_targets(y_train, y_test)
4 le.fit(y_train)
5 y_train_enc = le.transform(y_train)
—-> 6 y_test_enc = le.transform(y_test)
7 return y_train_enc, y_test_enc
~\anaconda3\envs\tensorflow\lib\site-packages\sklearn\preprocessing\_label.py in transform(self, y)
136 return np.array([])
137
–> 138 return _encode(y, uniques=self.classes_)
139
140 def inverse_transform(self, y):
~\anaconda3\envs\tensorflow\lib\site-packages\sklearn\utils\_encode.py in _encode(values, uniques, check_unknown)
183 diff = _check_unknown(values, uniques)
184 if diff:
–> 185 raise ValueError(f”y contains previously unseen labels: ”
186 f”{str(diff)}”)
187 return np.searchsorted(uniques, values)
ValueError: y contains previously unseen labels: [-0.0028116, -0.0019271, -0.001851, -0.0018209, -0.0018169, -0.0017662,-0.0016889, -0.0016376, -0.0016301, -0.0015716, -0.0015441, -0.0015289, -0.0015246, -0.001476, -0.0014491, -0.0014442, -0.0014414, -0.0014363, -0.0014354, -0.0014251, -0.0014239, -0.0013996, -0.0013764, -0.0013751, -0.0013727, -0.001372, -0.0013707, -0.0013691, -0.0013511, -0.001348, -0.0013289, -0.0013254, -0.0013088, -0.0012943, -0.0012932, -0.0012914, -0.0012854, -0.0012787, -0.0012716, -0.0012698, -0.0012695, -0.0012576, ……]
Seems to me you pass in numerical data into label transformer. May be you should check your code.
hi there, is it possible to transform the data first using ordinalencoding and labelencoding, then split it into training and testing set? thank you in advance
Yes. However, any transformation of this kind need to be careful not to leak the data.
what do you mean by ‘not to leak the data’ ?
It means not to make the output be part of the input and not to mix test and training set to create your preprocessing pipeline. No hint of the output should *leak* to the input.
i dont get it…but thank you
Maybe one example can help: All training examples are negative numbers and all test examples are positive numbers. You do min-max scaling with both, so your scaled data are in 0 to 1 with all training in 0 to 0.5 and all test in 0.5 to 1. Then my trained regression model will predict 0.75 and get a good result.
ooh so the prediction is not “learned” properly in training?
No, the prediction learned something it should not learned.
i see, thank you so much mr. adrian tam!
I can create a model by label-encoding the variables as shown above.
Next, I will create a model by OneHotEncoding the objective variable.
In the case of variables, they need to match the output layer, and in the case of OneHotEncoding, the values are arrays, so the fitting method will generate an error (
Failed to find data adapter that can handle input).Is it possible to create a model with OneHotEncoding for the objective variable?
Hi, thanks for the nice explanation!
I’m currently working on a time series prediction and want to use the explained embedding method for transforming my categorical features. Should I split the sequence before or after transforming the features to embeddings? I’m having a hard time figuring it out on my own…
Thanks a lot!
Hi Katharina…The following discussion may help add clarity:
https://datascience.stackexchange.com/questions/54908/data-normalization-before-or-after-train-test-split
Hi Jason,
With this model architecture and the X values get converted into list, how do you do cross validation or hyperparameter tuning using randomized or grid search?
Hi Tom…You may find the following of interest:
https://dev.to/balapriya/cross-validation-and-hyperparameter-search-in-scikit-learn-a-complete-guide-5ed8
TypeError: sparse matrix length is ambiguous; use getnnz() or shape[0]. Plz help me out
Sir plz guide me out regarding this error. Thanks.
TypeError: sparse matrix length is ambiguous; use getnnz() or shape[0]. Sir plz guide me out regarding this error. Thanks.
Hi Menahil…You may find the following of interest:
https://stackoverflow.com/questions/28314337/typeerror-sparse-matrix-length-is-ambiguous-use-getnnz-or-shape0-while-usi
Hi,
To me, it seems that the “closeness” of words is totally lost when the variables are label encoded. Please enlighten me..!
Thanks a lot for the nice article.
Hi Ryan…You are very welcome! Please elaborate on what you are seeking to accomplish that the concepts that were presented in the tutorial are not sufficient. That will better enable us to assist you.
For example,
a = [‘acid1’, ‘acid2’, ‘acid3’, ‘acid4’, ‘gas1’, ‘gas2’, ‘gas3’]
will be encoded like this:
a_enc = [0, 1, 2, 3, 4, 5, 6]
‘acid4’ and ‘gas1’ are close when encoded to 3 and 4, but totally unrelated. Shouldn’t the embeddings be derived directly from the strings, so that the resulting vectors for ‘acid4’ and ‘gas1’ are significantly different?
merge layer no longer exists. Please update
Hi! Thank you for this great articles and your other contributions.
I wanted to know your suggestions on how to encode if there is a categorical column that can be further divided into sub-categories. say – 11 categories(food, fashion, weather…) which are further divided into (continental, south-Indian, Punjabi: Food, dress, skirt, shoes: Fashion, sunny, windy: Weather).
Here the sub-categories and categories have some relation b/w them.
Hi Daneshwari…I would recommend that you further break down the classifications and apply multivariable classification:
https://machinelearningmastery.com/how-to-develop-rnn-models-for-human-activity-recognition-time-series-classification/
… More precisely encoding both categorical, subcategorical and their relation
… More precisely encoding both categorical, subcategory and their relation
When applying the embedding to new test data, do we need to keep and apply the label encoder(s) we fit during training?
Hi CC…More information regarding encoding can be found here:
https://machinelearningmastery.com/one-hot-encoding-for-categorical-data/
What if I have several categorical variables and I want to create embeddings for each one?
And I want to use them to fit a prediction model or a simple regression.
Is it a problem if the embedding outcomes of one variables are similar to the others?
Or are they independent?