Machine learning predictive modeling performance is only as good as your data, and your data is only as good as the way you prepare it for modeling.
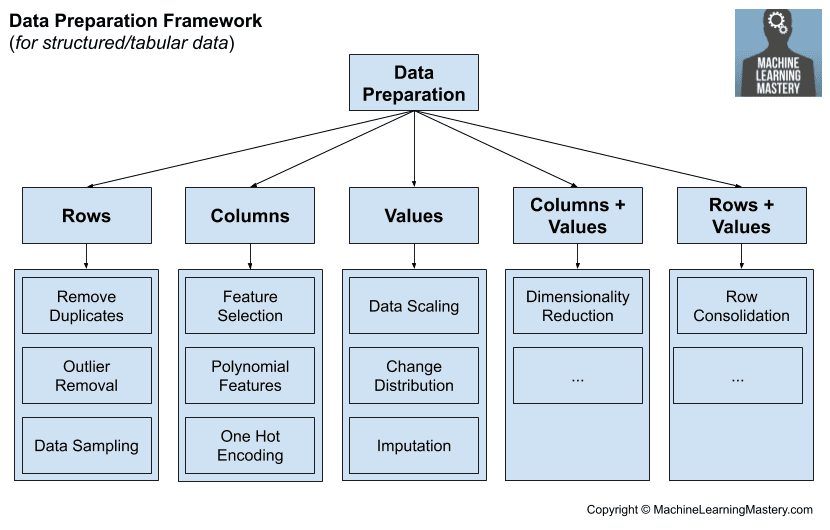
The most common approach to data preparation is to study a dataset and review the expectations of a machine learning algorithms, then carefully choose the most appropriate data preparation techniques to transform the raw data to best meet the expectations of the algorithm. This is slow, expensive, and requires a vast amount of expertise.
An alternative approach to data preparation is to grid search a suite of common and commonly useful data preparation techniques to the raw data. This is an alternative philosophy for data preparation that treats data transforms as another hyperparameter of the modeling pipeline to be searched and tuned.
This approach requires less expertise than the traditional manual approach to data preparation, although it is computationally costly. The benefit is that it can aid in the discovery of non-intuitive data preparation solutions that achieve good or best performance for a given predictive modeling problem.
In this tutorial, you will discover how to use the grid search approach for data preparation with tabular data.
After completing this tutorial, you will know:
- Grid search provides an alternative approach to data preparation for tabular data, where transforms are tried as hyperparameters of the modeling pipeline.
- How to use the grid search method for data preparation to improve model performance over a baseline for a standard classification dataset.
- How to grid search sequences of data preparation methods to further improve model performance.
Kick-start your project with my new book Data Preparation for Machine Learning, including step-by-step tutorials and the Python source code files for all examples.
Let’s get started.

How to Grid Search Data Preparation Techniques
Photo by Wall Boat, some rights reserved.
Tutorial Overview
This tutorial is divided into three parts; they are:
- Grid Search Technique for Data Preparation
- Dataset and Performance Baseline
- Wine Classification Dataset
- Baseline Model Performance
- Grid Search Approach to Data Preparation
Grid Search Technique for Data Preparation
Data preparation can be challenging.
The approach that is most often prescribed and followed is to analyze the dataset, review the requirements of the algorithms, and transform the raw data to best meet the expectations of the algorithms.
This can be effective but is also slow and can require deep expertise with data analysis and machine learning algorithms.
An alternative approach is to treat the preparation of input variables as a hyperparameter of the modeling pipeline and to tune it along with the choice of algorithm and algorithm configurations.
This might be a data transform that “should not work” or “should not be appropriate for the algorithm” yet results in good or great performance. Alternatively, it may be the absence of a data transform for an input variable that is deemed “absolutely required” yet results in good or great performance.
This can be achieved by designing a grid search of data preparation techniques and/or sequences of data preparation techniques in pipelines. This may involve evaluating each on a single chosen machine learning algorithm, or on a suite of machine learning algorithms.
The benefit of this approach is that it always results in suggestions of modeling pipelines that give good relative results. Most importantly, it can unearth the non-obvious and unintuitive solutions to practitioners without the need for deep expertise.
We can explore this approach to data preparation with a worked example.
Before we dive into a worked example, let’s first select a standard dataset and develop a baseline in performance.
Want to Get Started With Data Preparation?
Take my free 7-day email crash course now (with sample code).
Click to sign-up and also get a free PDF Ebook version of the course.
Dataset and Performance Baseline
In this section, we will first select a standard machine learning dataset and establish a baseline in performance on this dataset. This will provide the context for exploring the grid search method of data preparation in the next section.
Wine Classification Dataset
We will use the wine classification dataset.
This dataset has 13 input variables that describe the chemical composition of samples of wine and requires that the wine be classified as one of three types.
You can learn more about the dataset here:
No need to download the dataset as we will download it automatically as part of our worked examples.
Open the dataset and review the raw data. The first few rows of data are listed below.
We can see that it is a multi-class classification predictive modeling problem with numerical input variables, each of which has different scales.
|
1 2 3 4 5 6 |
14.23,1.71,2.43,15.6,127,2.8,3.06,.28,2.29,5.64,1.04,3.92,1065,1 13.2,1.78,2.14,11.2,100,2.65,2.76,.26,1.28,4.38,1.05,3.4,1050,1 13.16,2.36,2.67,18.6,101,2.8,3.24,.3,2.81,5.68,1.03,3.17,1185,1 14.37,1.95,2.5,16.8,113,3.85,3.49,.24,2.18,7.8,.86,3.45,1480,1 13.24,2.59,2.87,21,118,2.8,2.69,.39,1.82,4.32,1.04,2.93,735,1 ... |
The example below loads the dataset and splits it into the input and output columns, then summarizes the data arrays.
|
1 2 3 4 5 6 7 8 9 10 11 12 |
# example of loading and summarizing the wine dataset from pandas import read_csv # define the location of the dataset url = 'https://raw.githubusercontent.com/jbrownlee/Datasets/master/wine.csv' # load the dataset as a data frame df = read_csv(url, header=None) # retrieve the numpy array data = df.values # split the columns into input and output variables X, y = data[:, :-1], data[:, -1] # summarize the shape of the loaded data print(X.shape, y.shape) |
Running the example, we can see that the dataset was loaded correctly and that there are 179 rows of data with 13 input variables and a single target variable.
|
1 |
(178, 13) (178,) |
Next, let’s evaluate a model on this dataset and establish a baseline in performance.
Baseline Model Performance
We can establish a baseline in performance on the wine classification task by evaluating a model on the raw input data.
In this case, we will evaluate a logistic regression model.
First, we can define a function to load the dataset and perform some minimal data preparation to ensure the inputs are numeric and the target is label encoded.
|
1 2 3 4 5 6 7 8 9 10 11 |
# prepare the dataset def load_dataset(): # load the dataset url = 'https://raw.githubusercontent.com/jbrownlee/Datasets/master/wine.csv' df = read_csv(url, header=None) data = df.values X, y = data[:, :-1], data[:, -1] # minimally prepare dataset X = X.astype('float') y = LabelEncoder().fit_transform(y.astype('str')) return X, y |
We will evaluate the model using the gold standard of repeated stratified k-fold cross-validation with 10 folds and three repeats.
|
1 2 3 4 5 6 7 |
# evaluate a model def evaluate_model(X, y, model): # define the cross-validation procedure cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1) # evaluate model scores = cross_val_score(model, X, y, scoring='accuracy', cv=cv, n_jobs=-1) return scores |
We can then call the function to load the dataset, define our model, then evaluate it, reporting the mean and standard deviation accuracy.
|
1 2 3 4 5 6 7 8 9 |
... # get the dataset X, y = load_dataset() # define the model model = LogisticRegression(solver='liblinear') # evaluate the model scores = evaluate_model(X, y, model) # report performance print('Accuracy: %.3f (%.3f)' % (mean(scores), std(scores))) |
Tying this together, the complete example of evaluating a logistic regression model on the raw wine classification dataset is listed below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 |
# baseline model performance on the wine dataset from numpy import mean from numpy import std from pandas import read_csv from sklearn.preprocessing import LabelEncoder from sklearn.model_selection import RepeatedStratifiedKFold from sklearn.model_selection import cross_val_score from sklearn.linear_model import LogisticRegression # prepare the dataset def load_dataset(): # load the dataset url = 'https://raw.githubusercontent.com/jbrownlee/Datasets/master/wine.csv' df = read_csv(url, header=None) data = df.values X, y = data[:, :-1], data[:, -1] # minimally prepare dataset X = X.astype('float') y = LabelEncoder().fit_transform(y.astype('str')) return X, y # evaluate a model def evaluate_model(X, y, model): # define the cross-validation procedure cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1) # evaluate model scores = cross_val_score(model, X, y, scoring='accuracy', cv=cv, n_jobs=-1) return scores # get the dataset X, y = load_dataset() # define the model model = LogisticRegression(solver='liblinear') # evaluate the model scores = evaluate_model(X, y, model) # report performance print('Accuracy: %.3f (%.3f)' % (mean(scores), std(scores))) |
Running the example evaluates the model performance and reports the mean and standard deviation classification accuracy.
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
In this case, we can see that the logistic regression model fit on the raw input data achieved the average classification accuracy of about 95.3 percent, providing a baseline in performance.
|
1 |
Accuracy: 0.953 (0.048) |
Next, let’s explore whether we can improve the performance using the grid-search-based approach to data preparation.
Grid Search Approach to Data Preparation
In this section, we can explore whether we can improve performance using the grid search approach to data preparation.
The first step is to define a series of modeling pipelines to evaluate, where each pipeline defines one (or more) data preparation techniques and ends with a model that takes the transformed data as input.
We will define a function to create these pipelines as a list of tuples, where each tuple defines the short name for the pipeline and the pipeline itself. We will evaluate a range of different data scaling methods (e.g. MinMaxScaler and StandardScaler), distribution transforms (QuantileTransformer and KBinsDiscretizer), as well as dimensionality reduction transforms (PCA and SVD).
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 |
# get modeling pipelines to evaluate def get_pipelines(model): pipelines = list() # normalize p = Pipeline([('s',MinMaxScaler()), ('m',model)]) pipelines.append(('norm', p)) # standardize p = Pipeline([('s',StandardScaler()), ('m',model)]) pipelines.append(('std', p)) # quantile p = Pipeline([('s',QuantileTransformer(n_quantiles=100, output_distribution='normal')), ('m',model)]) pipelines.append(('quan', p)) # discretize p = Pipeline([('s',KBinsDiscretizer(n_bins=10, encode='ordinal', strategy='uniform')), ('m',model)]) pipelines.append(('kbins', p)) # pca p = Pipeline([('s',PCA(n_components=7)), ('m',model)]) pipelines.append(('pca', p)) # svd p = Pipeline([('s',TruncatedSVD(n_components=7)), ('m',model)]) pipelines.append(('svd', p)) return pipelines |
We can then call this function to get the list of transforms, then enumerate each, evaluating it and reporting the performance along the way.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 |
... # get the modeling pipelines pipelines = get_pipelines(model) # evaluate each pipeline results, names = list(), list() for name, pipeline in pipelines: # evaluate scores = evaluate_model(X, y, pipeline) # summarize print('>%s: %.3f (%.3f)' % (name, mean(scores), std(scores))) # store results.append(scores) names.append(name) |
At the end of the run, we can create a box and whisker plot for each set of scores and compare the distributions of results visually.
|
1 2 3 4 |
... # plot the result pyplot.boxplot(results, labels=names, showmeans=True) pyplot.show() |
Tying this together, the complete example of grid searching data preparation techniques on the wine classification dataset is listed below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 |
# compare data preparation methods for the wine classification dataset from numpy import mean from numpy import std from pandas import read_csv from sklearn.model_selection import RepeatedStratifiedKFold from sklearn.model_selection import cross_val_score from sklearn.linear_model import LogisticRegression from sklearn.pipeline import Pipeline from sklearn.preprocessing import LabelEncoder from sklearn.preprocessing import MinMaxScaler from sklearn.preprocessing import StandardScaler from sklearn.preprocessing import QuantileTransformer from sklearn.preprocessing import KBinsDiscretizer from sklearn.decomposition import PCA from sklearn.decomposition import TruncatedSVD from matplotlib import pyplot # prepare the dataset def load_dataset(): # load the dataset url = 'https://raw.githubusercontent.com/jbrownlee/Datasets/master/wine.csv' df = read_csv(url, header=None) data = df.values X, y = data[:, :-1], data[:, -1] # minimally prepare dataset X = X.astype('float') y = LabelEncoder().fit_transform(y.astype('str')) return X, y # evaluate a model def evaluate_model(X, y, model): # define the cross-validation procedure cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1) # evaluate model scores = cross_val_score(model, X, y, scoring='accuracy', cv=cv, n_jobs=-1) return scores # get modeling pipelines to evaluate def get_pipelines(model): pipelines = list() # normalize p = Pipeline([('s',MinMaxScaler()), ('m',model)]) pipelines.append(('norm', p)) # standardize p = Pipeline([('s',StandardScaler()), ('m',model)]) pipelines.append(('std', p)) # quantile p = Pipeline([('s',QuantileTransformer(n_quantiles=100, output_distribution='normal')), ('m',model)]) pipelines.append(('quan', p)) # discretize p = Pipeline([('s',KBinsDiscretizer(n_bins=10, encode='ordinal', strategy='uniform')), ('m',model)]) pipelines.append(('kbins', p)) # pca p = Pipeline([('s',PCA(n_components=7)), ('m',model)]) pipelines.append(('pca', p)) # svd p = Pipeline([('s',TruncatedSVD(n_components=7)), ('m',model)]) pipelines.append(('svd', p)) return pipelines # get the dataset X, y = load_dataset() # define the model model = LogisticRegression(solver='liblinear') # get the modeling pipelines pipelines = get_pipelines(model) # evaluate each pipeline results, names = list(), list() for name, pipeline in pipelines: # evaluate scores = evaluate_model(X, y, pipeline) # summarize print('>%s: %.3f (%.3f)' % (name, mean(scores), std(scores))) # store results.append(scores) names.append(name) # plot the result pyplot.boxplot(results, labels=names, showmeans=True) pyplot.show() |
Running the example evaluates the performance of each pipeline and reports the mean and standard deviation classification accuracy.
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
In this case, we can see that standardizing the input variables and using a quantile transform both achieves the best result with a classification accuracy of about 98.7 percent, an improvement over the baseline with no data preparation that achieved a classification accuracy of 95.3 percent.
You can add your own modeling pipelines to the get_pipelines() function and compare their result.
Can you get better results?
Let me know in the comments below.
|
1 2 3 4 5 6 |
>norm: 0.976 (0.031) >std: 0.987 (0.023) >quan: 0.987 (0.023) >kbins: 0.968 (0.045) >pca: 0.963 (0.039) >svd: 0.953 (0.048) |
A figure is created showing box and whisker plots that summarize the distribution of classification accuracy scores for each data preparation technique. We can see that the distribution of scores for the standardization and quantile transforms are compact and very similar and have an outlier. We can see that the spread of scores for the other transforms is larger and skewing down.
The results may suggest that standardizing the dataset is probably an important step in the data preparation and related transforms, such as the quantile transform, and perhaps even the power transform may offer benefits if combined with standardization by making one or more input variables more Gaussian.
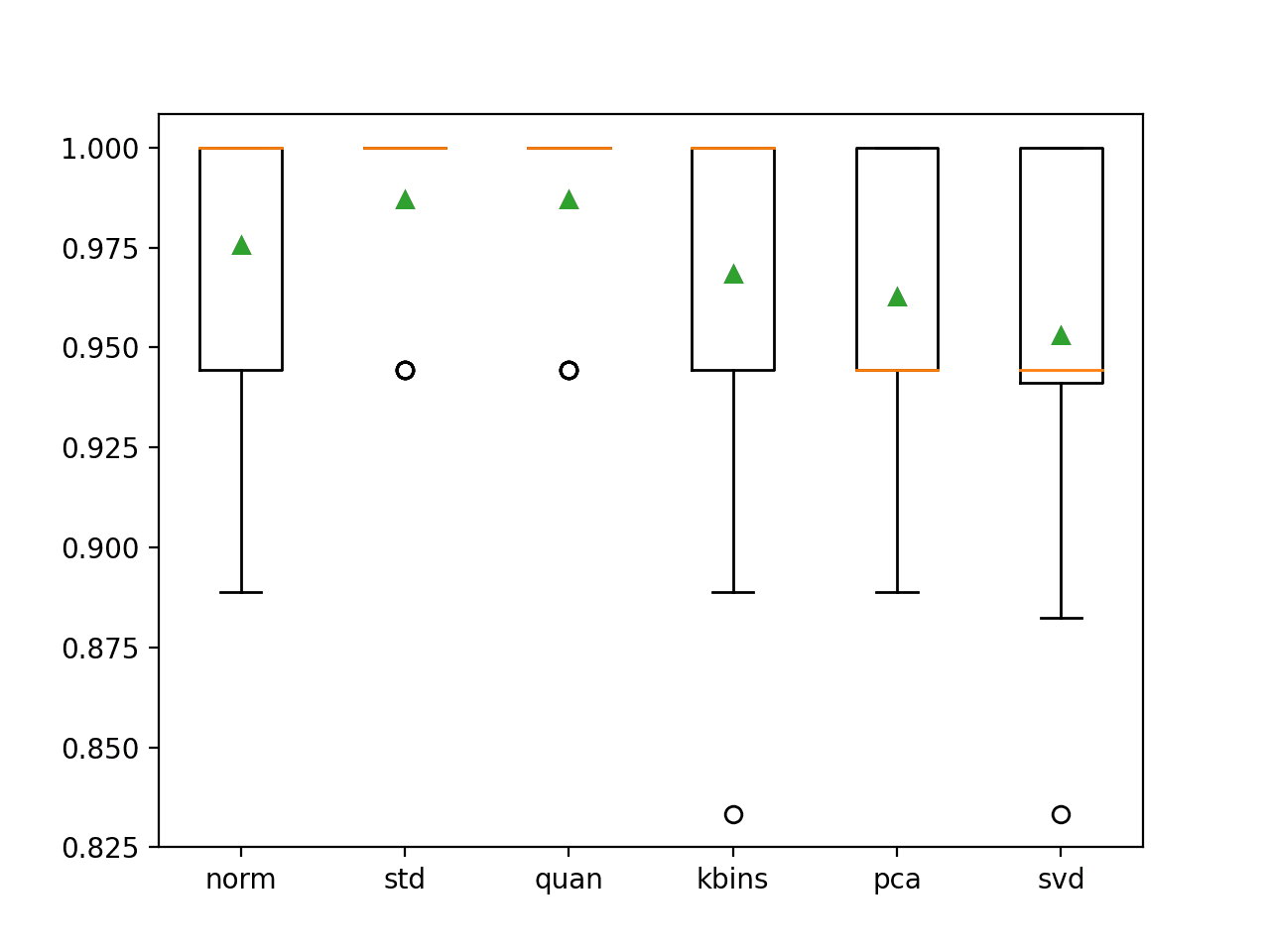
Box and Whisker Plot of Classification Accuracy for Different Data Transforms on the Wine Classification Dataset
We can also explore sequences of transforms to see if they can offer a lift in performance.
For example, we might want to apply RFE feature selection after the standardization transform to see if the same or better results can be used with fewer input variables (e.g. less complexity).
We might also want to see if a power transform preceded with a data scaling transform can achieve good performance on the dataset as we believe it could given the success of the quantile transform.
The updated get_pipelines() function with sequences of transforms is provided below.
|
1 2 3 4 5 6 7 8 9 10 |
# get modeling pipelines to evaluate def get_pipelines(model): pipelines = list() # standardize p = Pipeline([('s',StandardScaler()), ('r', RFE(estimator=LogisticRegression(solver='liblinear'), n_features_to_select=10)), ('m',model)]) pipelines.append(('std', p)) # scale and power p = Pipeline([('s',MinMaxScaler((1,2))), ('p', PowerTransformer()), ('m',model)]) pipelines.append(('power', p)) return pipelines |
Tying this together, the complete example is listed below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 |
# compare sequences of data preparation methods for the wine classification dataset from numpy import mean from numpy import std from pandas import read_csv from sklearn.model_selection import RepeatedStratifiedKFold from sklearn.model_selection import cross_val_score from sklearn.linear_model import LogisticRegression from sklearn.pipeline import Pipeline from sklearn.preprocessing import LabelEncoder from sklearn.preprocessing import MinMaxScaler from sklearn.preprocessing import StandardScaler from sklearn.preprocessing import QuantileTransformer from sklearn.preprocessing import PowerTransformer from sklearn.preprocessing import KBinsDiscretizer from sklearn.decomposition import PCA from sklearn.decomposition import TruncatedSVD from sklearn.feature_selection import RFE from matplotlib import pyplot # prepare the dataset def load_dataset(): # load the dataset url = 'https://raw.githubusercontent.com/jbrownlee/Datasets/master/wine.csv' df = read_csv(url, header=None) data = df.values X, y = data[:, :-1], data[:, -1] # minimally prepare dataset X = X.astype('float') y = LabelEncoder().fit_transform(y.astype('str')) return X, y # evaluate a model def evaluate_model(X, y, model): # define the cross-validation procedure cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1) # evaluate model scores = cross_val_score(model, X, y, scoring='accuracy', cv=cv, n_jobs=-1) return scores # get modeling pipelines to evaluate def get_pipelines(model): pipelines = list() # standardize p = Pipeline([('s',StandardScaler()), ('r', RFE(estimator=LogisticRegression(solver='liblinear'), n_features_to_select=10)), ('m',model)]) pipelines.append(('std', p)) # scale and power p = Pipeline([('s',MinMaxScaler((1,2))), ('p', PowerTransformer()), ('m',model)]) pipelines.append(('power', p)) return pipelines # get the dataset X, y = load_dataset() # define the model model = LogisticRegression(solver='liblinear') # get the modeling pipelines pipelines = get_pipelines(model) # evaluate each pipeline results, names = list(), list() for name, pipeline in pipelines: # evaluate scores = evaluate_model(X, y, pipeline) # summarize print('>%s: %.3f (%.3f)' % (name, mean(scores), std(scores))) # store results.append(scores) names.append(name) # plot the result pyplot.boxplot(results, labels=names, showmeans=True) pyplot.show() |
Running the example evaluates the performance of each pipeline and reports the mean and standard deviation classification accuracy.
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
In this case, we can see that the standardization with feature selection offers an additional lift in accuracy from 98.7 percent to 98.9 percent, although the data scaling and power transform do not offer any additional benefit over the quantile transform.
|
1 2 |
>std: 0.989 (0.022) >power: 0.987 (0.023) |
A figure is created showing box and whisker plots that summarize the distribution of classification accuracy scores for each data preparation technique.
We can see that the distribution of results for both pipelines of transforms is compact with very little spread other than outlier.
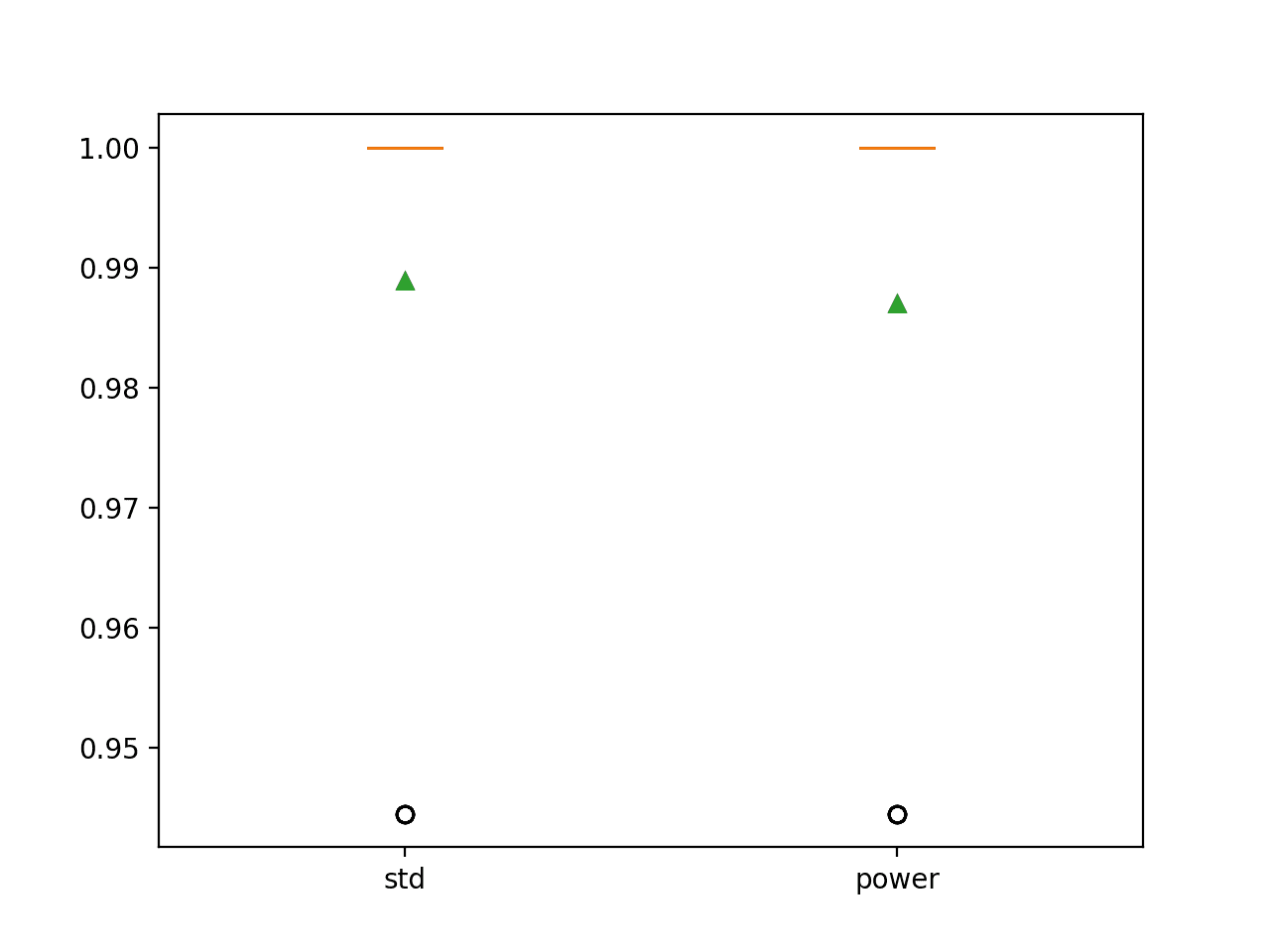
Box and Whisker Plot of Classification Accuracy for Different Sequences of Data Transforms on the Wine Classification Dataset
Further Reading
This section provides more resources on the topic if you are looking to go deeper.
Books
APIs
Summary
In this tutorial, you discovered how to use a grid search approach for data preparation with tabular data.
Specifically, you learned:
- Grid search provides an alternative approach to data preparation for tabular data, where transforms are tried as hyperparameters of the modeling pipeline.
- How to use the grid search method for data preparation to improve model performance over a baseline for a standard classification dataset.
- How to grid search sequences of data preparation methods to further improve model performance.
Do you have any questions?
Ask your questions in the comments below and I will do my best to answer.





Nice post as always. May i know how to apply grid search for LSTM (neural network) model? Bcoz its very important to choose the hyper parameters for best results. Thanks in advance jason
Yes, see this tutorial:
https://machinelearningmastery.com/how-to-grid-search-deep-learning-models-for-time-series-forecasting/
Dear Dr Jason,
I downloaded the file ‘wine.csv’ from your site.
There seems to be a format error when loading the dataset.
If I downloaded as in the above code:
The above does not look right. So I added another parameter in read_csv
Conclusion
For the downloaded file wine.csv, you need to have read_csv include the parameter delim_whitespace = True, otherwise without the parameter, you get the mess shown at the beginning of my comment
Hope that helps,
Anthony of Sydney
Here is the dataset in CSV format:
https://raw.githubusercontent.com/jbrownlee/Datasets/master/wine.csv
All datasets are located here:
https://github.com/jbrownlee/Datasets
Dear Dr Jason,
Thank you for this tutorial.
Under the subheading “Grid Search Approach to Data Preparation” there is a function called
I noticed that passing the ‘model’ parameter did not get attached to the pipeline. Nor was the ‘model’ added as part of a pipeline.
So I re-wrote the module without passing the ‘model’ parameter such as:
I obtained the same result using the get_pipelines() function.
What is the purpose of passing the ‘model’ variable in get_pipelines? It does not get attached to a particular pipeline within the get_pipelines(model) function.
Not including ‘model’ parameter in the get_pipelines() function produced the same result
Thank you,
Anthony of Sydney.
Dear Dr Jason,
PROBLEM SOLVED – THE ‘model’ variable is still used as a step for various pipelines within get_pipelines() because it ‘model’ is a global variable.
The reason why we got the same result is that whether get_pipelines(model) OR get_pipelines(), the ‘model’ is still added to the steps within the get_pipelines(model) or get_pipelines().
How does model get passed without needing the ‘model’ parameter in get_pipelines()?
Answer is that ‘model’ is GLOBAL.
How is that GLOBAL?
Answer ‘model’ is a global variable which the function get_pipelines() uses the globally declared variable ‘model’ within the get_pipelines().
You may ask how does it get used within get_pipelines()?
Answer: here is a typical few lines used in get_pipelines():
Conclusion – this is the danger of using global variables. If a module/function needs to be passed to a module, the parameter needs to be a different name to the global variable to limit the scope of the global variable.
Thank you,
Anthony of Sydney
No, “model” is passed as an argument to get_pipelines() where it is added to the end of each pipeline.
The model is added to the end of each pipeline in that function.
Dear Dr Jason,
Thank you for your reply. I should have been clearer.
Yes ‘model’ is added to the end of each pipeline “,,,within that function.” At the same time, there was no need to declare the function as get_pipelines(model) since ‘model’ was (i) declared outside the function and (ii) because ‘model’ was declared outside the function, ‘model’ was able to be used within get_pipelines() without passing the parameter ‘model’ in your version of get_pipelines(model).
Thus ‘model’ in your example is global.
Thank you, your response is appreciated,
Anthony of Sydney
Yes, but I don’t recommend using/relying-on globals, therefore it was provided as an argument to the function for use within the function.
Dear Dr Jason,
Thank you, it is appreciated. It clarified the danger of global variables.
Anthony of Sydney.