In this tutorial, you will discover how to implement the Bayesian Optimization algorithm for complex optimization problems.
Global optimization is a challenging problem of finding an input that results in the minimum or maximum cost of a given objective function.
Typically, the form of the objective function is complex and intractable to analyze and is often non-convex, nonlinear, high dimension, noisy, and computationally expensive to evaluate.
Bayesian Optimization provides a principled technique based on Bayes Theorem to direct a search of a global optimization problem that is efficient and effective. It works by building a probabilistic model of the objective function, called the surrogate function, that is then searched efficiently with an acquisition function before candidate samples are chosen for evaluation on the real objective function.
Bayesian Optimization is often used in applied machine learning to tune the hyperparameters of a given well-performing model on a validation dataset.
After completing this tutorial, you will know:
- Global optimization is a challenging problem that involves black box and often non-convex, non-linear, noisy, and computationally expensive objective functions.
- Bayesian Optimization provides a probabilistically principled method for global optimization.
- How to implement Bayesian Optimization from scratch and how to use open-source implementations.
Kick-start your project with my new book Probability for Machine Learning, including step-by-step tutorials and the Python source code files for all examples.
Let’s get started.
- Update Jan/2020: Updated for changes in scikit-learn v0.22 API.

A Gentle Introduction to Bayesian Optimization
Photo by Beni Arnold, some rights reserved.
Tutorial Overview
This tutorial is divided into four parts; they are:
- Challenge of Function Optimization
- What Is Bayesian Optimization
- How to Perform Bayesian Optimization
- Hyperparameter Tuning With Bayesian Optimization
Challenge of Function Optimization
Global function optimization, or function optimization for short, involves finding the minimum or maximum of an objective function.
Samples are drawn from the domain and evaluated by the objective function to give a score or cost.
Let’s define some common terms:
- Samples. One example from the domain, represented as a vector.
- Search Space: Extent of the domain from which samples can be drawn.
- Objective Function. Function that takes a sample and returns a cost.
- Cost. Numeric score for a sample calculated via the objective function.
Samples are comprised of one or more variables generally easy to devise or create. One sample is often defined as a vector of variables with a predefined range in an n-dimensional space. This space must be sampled and explored in order to find the specific combination of variable values that result in the best cost.
The cost often has units that are specific to a given domain. Optimization is often described in terms of minimizing cost, as a maximization problem can easily be transformed into a minimization problem by inverting the calculated cost. Together, the minimum and maximum of a function are referred to as the extreme of the function (or the plural extrema).
The objective function is often easy to specify but can be computationally challenging to calculate or result in a noisy calculation of cost over time. The form of the objective function is unknown and is often highly nonlinear, and highly multi-dimensional defined by the number of input variables. The function is also probably non-convex. This means that local extrema may or may not be the global extrema (e.g. could be misleading and result in premature convergence), hence the name of the task as global rather than local optimization.
Although little is known about the objective function, (it is known whether the minimum or the maximum cost from the function is sought), and as such, it is often referred to as a black box function and the search process as black box optimization. Further, the objective function is sometimes called an oracle given the ability to only give answers.
Function optimization is a fundamental part of machine learning. Most machine learning algorithms involve the optimization of parameters (weights, coefficients, etc.) in response to training data. Optimization also refers to the process of finding the best set of hyperparameters that configure the training of a machine learning algorithm. Taking one step higher again, the selection of training data, data preparation, and machine learning algorithms themselves is also a problem of function optimization.
Summary of optimization in machine learning:
- Algorithm Training. Optimization of model parameters.
- Algorithm Tuning. Optimization of model hyperparameters.
- Predictive Modeling. Optimization of data, data preparation, and algorithm selection.
Many methods exist for function optimization, such as randomly sampling the variable search space, called random search, or systematically evaluating samples in a grid across the search space, called grid search.
More principled methods are able to learn from sampling the space so that future samples are directed toward the parts of the search space that are most likely to contain the extrema.
A directed approach to global optimization that uses probability is called Bayesian Optimization.
Want to Learn Probability for Machine Learning
Take my free 7-day email crash course now (with sample code).
Click to sign-up and also get a free PDF Ebook version of the course.
What Is Bayesian Optimization
Bayesian Optimization is an approach that uses Bayes Theorem to direct the search in order to find the minimum or maximum of an objective function.
It is an approach that is most useful for objective functions that are complex, noisy, and/or expensive to evaluate.
Bayesian optimization is a powerful strategy for finding the extrema of objective functions that are expensive to evaluate. […] It is particularly useful when these evaluations are costly, when one does not have access to derivatives, or when the problem at hand is non-convex.
Recall that Bayes Theorem is an approach for calculating the conditional probability of an event:
- P(A|B) = P(B|A) * P(A) / P(B)
We can simplify this calculation by removing the normalizing value of P(B) and describe the conditional probability as a proportional quantity. This is useful as we are not interested in calculating a specific conditional probability, but instead in optimizing a quantity.
- P(A|B) = P(B|A) * P(A)
The conditional probability that we are calculating is referred to generally as the posterior probability; the reverse conditional probability is sometimes referred to as the likelihood, and the marginal probability is referred to as the prior probability; for example:
- posterior = likelihood * prior
This provides a framework that can be used to quantify the beliefs about an unknown objective function given samples from the domain and their evaluation via the objective function.
We can devise specific samples (x1, x2, …, xn) and evaluate them using the objective function f(xi) that returns the cost or outcome for the sample xi. Samples and their outcome are collected sequentially and define our data D, e.g. D = {xi, f(xi), … xn, f(xn)} and is used to define the prior. The likelihood function is defined as the probability of observing the data given the function P(D | f). This likelihood function will change as more observations are collected.
- P(f|D) = P(D|f) * P(f)
The posterior represents everything we know about the objective function. It is an approximation of the objective function and can be used to estimate the cost of different candidate samples that we may want to evaluate.
In this way, the posterior probability is a surrogate objective function.
The posterior captures the updated beliefs about the unknown objective function. One may also interpret this step of Bayesian optimization as estimating the objective function with a surrogate function (also called a response surface).
- Surrogate Function: Bayesian approximation of the objective function that can be sampled efficiently.
The surrogate function gives us an estimate of the objective function, which can be used to direct future sampling. Sampling involves careful use of the posterior in a function known as the “acquisition” function, e.g. for acquiring more samples. We want to use our belief about the objective function to sample the area of the search space that is most likely to pay off, therefore the acquisition will optimize the conditional probability of locations in the search to generate the next sample.
- Acquisition Function: Technique by which the posterior is used to select the next sample from the search space.
Once additional samples and their evaluation via the objective function f() have been collected, they are added to data D and the posterior is then updated.
This process is repeated until the extrema of the objective function is located, a good enough result is located, or resources are exhausted.
The Bayesian Optimization algorithm can be summarized as follows:
- 1. Select a Sample by Optimizing the Acquisition Function.
- 2. Evaluate the Sample With the Objective Function.
- 3. Update the Data and, in turn, the Surrogate Function.
- 4. Go To 1.
How to Perform Bayesian Optimization
In this section, we will explore how Bayesian Optimization works by developing an implementation from scratch for a simple one-dimensional test function.
First, we will define the test problem, then how to model the mapping of inputs to outputs with a surrogate function. Next, we will see how the surrogate function can be searched efficiently with an acquisition function before tying all of these elements together into the Bayesian Optimization procedure.
Test Problem
The first step is to define a test problem.
We will use a multimodal problem with five peaks, calculated as:
- y = x^2 * sin(5 * PI * x)^6
Where x is a real value in the range [0,1] and PI is the value of pi.
We will augment this function by adding Gaussian noise with a mean of zero and a standard deviation of 0.1. This will mean that the real evaluation will have a positive or negative random value added to it, making the function challenging to optimize.
The objective() function below implements this.
|
1 2 3 4 |
# objective function def objective(x, noise=0.1): noise = normal(loc=0, scale=noise) return (x**2 * sin(5 * pi * x)**6.0) + noise |
We can test this function by first defining a grid-based sample of inputs from 0 to 1 with a step size of 0.01 across the domain.
|
1 2 3 |
... # grid-based sample of the domain [0,1] X = arange(0, 1, 0.01) |
We can then evaluate these samples using the target function without any noise to see what the real objective function looks like.
|
1 2 3 |
... # sample the domain without noise y = [objective(x, 0) for x in X] |
We can then evaluate these same points with noise to see what the objective function will look like when we are optimizing it.
|
1 2 3 |
... # sample the domain with noise ynoise = [objective(x) for x in X] |
We can look at all of the non-noisy objective function values to find the input that resulted in the best score and report it. This will be the optima, in this case, maxima, as we are maximizing the output of the objective function.
We would not know this in practice, but for out test problem, it is good to know the real best input and output of the function to see if the Bayesian Optimization algorithm can locate it.
|
1 2 3 4 |
... # find best result ix = argmax(y) print('Optima: x=%.3f, y=%.3f' % (X[ix], y[ix])) |
Finally, we can create a plot, first showing the noisy evaluation as a scatter plot with input on the x-axis and score on the y-axis, then a line plot of the scores without any noise.
|
1 2 3 4 5 6 7 |
... # plot the points with noise pyplot.scatter(X, ynoise) # plot the points without noise pyplot.plot(X, y) # show the plot pyplot.show() |
The complete example of reviewing the test function that we wish to optimize is listed below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 |
# example of the test problem from math import sin from math import pi from numpy import arange from numpy import argmax from numpy.random import normal from matplotlib import pyplot # objective function def objective(x, noise=0.1): noise = normal(loc=0, scale=noise) return (x**2 * sin(5 * pi * x)**6.0) + noise # grid-based sample of the domain [0,1] X = arange(0, 1, 0.01) # sample the domain without noise y = [objective(x, 0) for x in X] # sample the domain with noise ynoise = [objective(x) for x in X] # find best result ix = argmax(y) print('Optima: x=%.3f, y=%.3f' % (X[ix], y[ix])) # plot the points with noise pyplot.scatter(X, ynoise) # plot the points without noise pyplot.plot(X, y) # show the plot pyplot.show() |
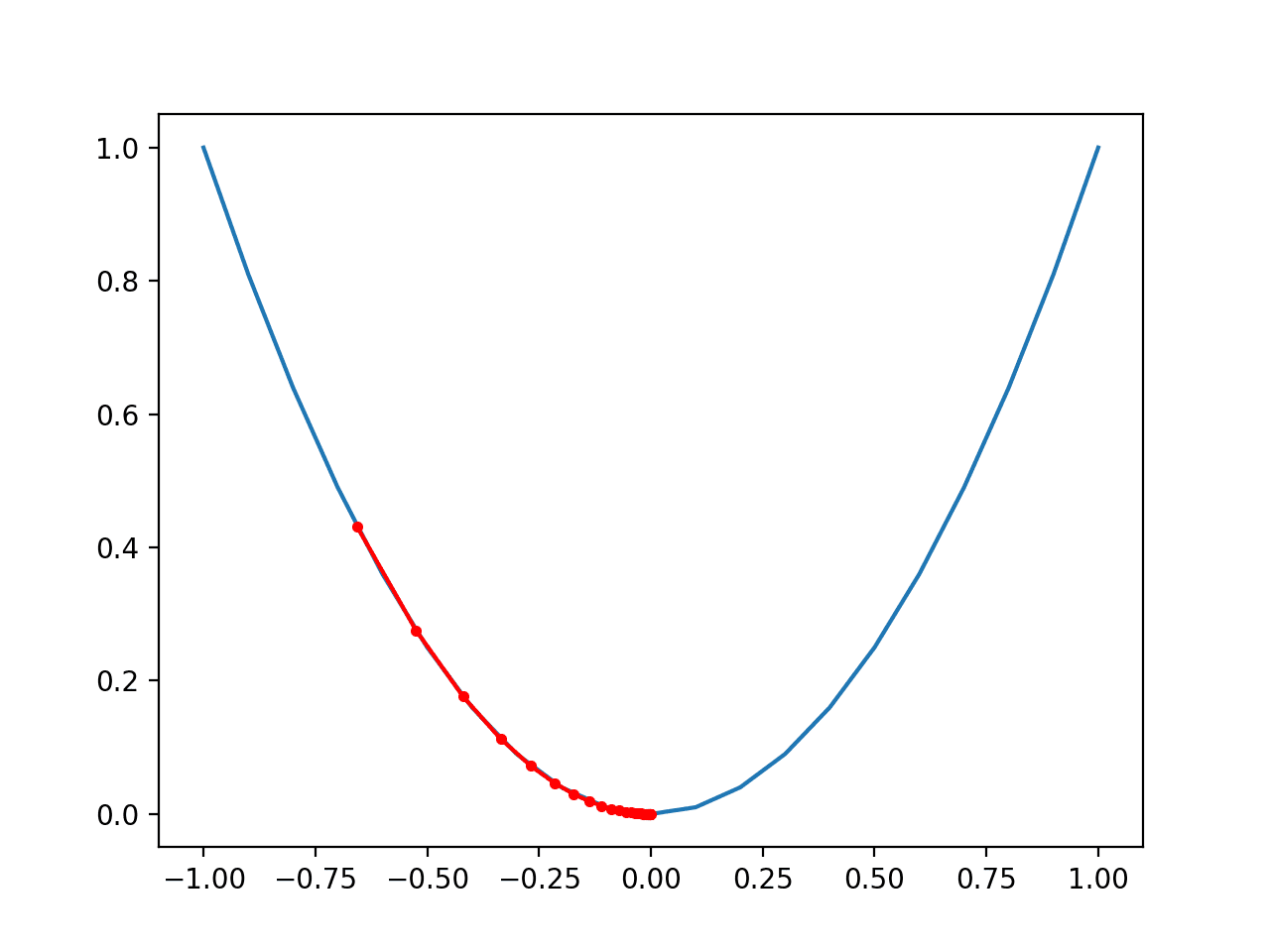
Running the example first reports the global optima as an input with the value 0.9 that gives the score 0.81.
|
1 |
Optima: x=0.900, y=0.810 |
A plot is then created showing the noisy evaluation of the samples (dots) and the non-noisy and true shape of the objective function (line).
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
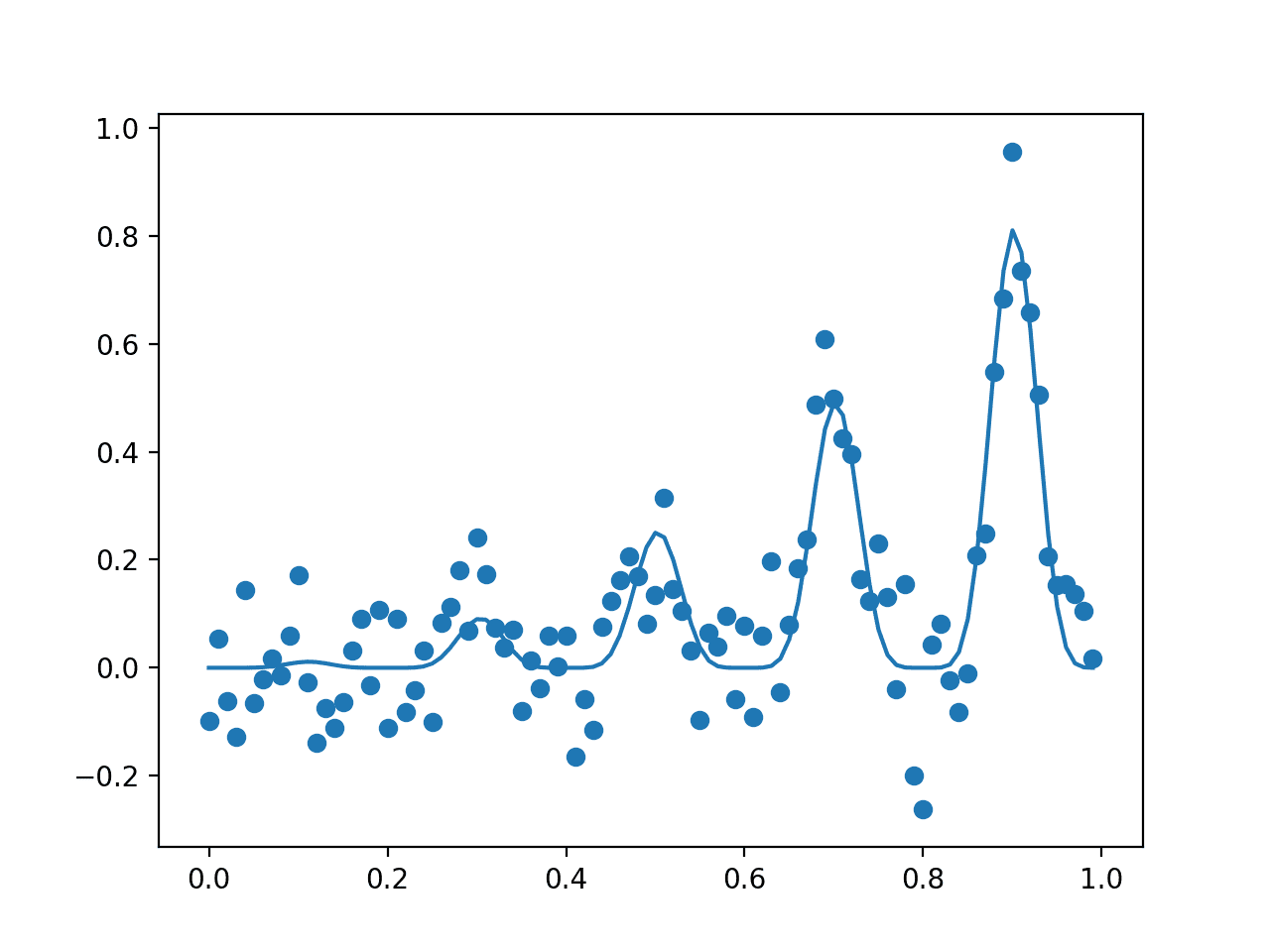
Plot of The Input Samples Evaluated with a Noisy (dots) and Non-Noisy (Line) Objective Function
Now that we have a test problem, let’s review how to train a surrogate function.
Surrogate Function
The surrogate function is a technique used to best approximate the mapping of input examples to an output score.
Probabilistically, it summarizes the conditional probability of an objective function (f), given the available data (D) or P(f|D).
A number of techniques can be used for this, although the most popular is to treat the problem as a regression predictive modeling problem with the data representing the input and the score representing the output to the model. This is often best modeled using a random forest or a Gaussian Process.
A Gaussian Process, or GP, is a model that constructs a joint probability distribution over the variables, assuming a multivariate Gaussian distribution. As such, it is capable of efficient and effective summarization of a large number of functions and smooth transition as more observations are made available to the model.
This smooth structure and smooth transition to new functions based on data are desirable properties as we sample the domain, and the multivariate Gaussian basis to the model means that an estimate from the model will be a mean of a distribution with a standard deviation; that will be helpful later in the acquisition function.
As such, using a GP regression model is often preferred.
We can fit a GP regression model using the GaussianProcessRegressor scikit-learn implementation from a sample of inputs (X) and noisy evaluations from the objective function (y).
First, the model must be defined. An important aspect in defining the GP model is the kernel. This controls the shape of the function at specific points based on distance measures between actual data observations. Many different kernel functions can be used, and some may offer better performance for specific datasets.
By default, a Radial Basis Function, or RBF, is used that can work well.
|
1 2 3 |
... # define the model model = GaussianProcessRegressor() |
Once defined, the model can be fit on the training dataset directly by calling the fit() function.
The defined model can be fit again at any time with updated data concatenated to the existing data by another call to fit().
|
1 2 3 |
... # fit the model model.fit(X, y) |
The model will estimate the cost for one or more samples provided to it.
The model is used by calling the predict() function. The result for a given sample will be a mean of the distribution at that point. We can also get the standard deviation of the distribution at that point in the function by specifying the argument return_std=True; for example:
|
1 2 |
... yhat = model.predict(X, return_std=True) |
This function can result in warnings if the distribution is thin at a given point we are interested in sampling.
Therefore, we can silence all of the warnings when making a prediction. The surrogate() function below takes the fit model and one or more samples and returns the mean and standard deviation estimated costs whilst not printing any warnings.
|
1 2 3 4 5 6 7 |
# surrogate or approximation for the objective function def surrogate(model, X): # catch any warning generated when making a prediction with catch_warnings(): # ignore generated warnings simplefilter("ignore") return model.predict(X, return_std=True) |
We can call this function any time to estimate the cost of one or more samples, such as when we want to optimize the acquisition function in the next section.
For now, it is interesting to see what the surrogate function looks like across the domain after it is trained on a random sample.
We can achieve this by first fitting the GP model on a random sample of 100 data points and their real objective function values with noise. We can then plot a scatter plot of these points. Next, we can perform a grid-based sample across the input domain and estimate the cost at each point using the surrogate function and plot the result as a line.
We would expect the surrogate function to have a crude approximation of the true non-noisy objective function.
The plot() function below creates this plot, given the random data sample of the real noisy objective function and the fit model.
|
1 2 3 4 5 6 7 8 9 10 11 |
# plot real observations vs surrogate function def plot(X, y, model): # scatter plot of inputs and real objective function pyplot.scatter(X, y) # line plot of surrogate function across domain Xsamples = asarray(arange(0, 1, 0.001)) Xsamples = Xsamples.reshape(len(Xsamples), 1) ysamples, _ = surrogate(model, Xsamples) pyplot.plot(Xsamples, ysamples) # show the plot pyplot.show() |
Tying this together, the complete example of fitting a Gaussian Process regression model on noisy samples and plotting the sample vs. the surrogate function is listed below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 |
# example of a gaussian process surrogate function from math import sin from math import pi from numpy import arange from numpy import asarray from numpy.random import normal from numpy.random import random from matplotlib import pyplot from warnings import catch_warnings from warnings import simplefilter from sklearn.gaussian_process import GaussianProcessRegressor # objective function def objective(x, noise=0.1): noise = normal(loc=0, scale=noise) return (x**2 * sin(5 * pi * x)**6.0) + noise # surrogate or approximation for the objective function def surrogate(model, X): # catch any warning generated when making a prediction with catch_warnings(): # ignore generated warnings simplefilter("ignore") return model.predict(X, return_std=True) # plot real observations vs surrogate function def plot(X, y, model): # scatter plot of inputs and real objective function pyplot.scatter(X, y) # line plot of surrogate function across domain Xsamples = asarray(arange(0, 1, 0.001)) Xsamples = Xsamples.reshape(len(Xsamples), 1) ysamples, _ = surrogate(model, Xsamples) pyplot.plot(Xsamples, ysamples) # show the plot pyplot.show() # sample the domain sparsely with noise X = random(100) y = asarray([objective(x) for x in X]) # reshape into rows and cols X = X.reshape(len(X), 1) y = y.reshape(len(y), 1) # define the model model = GaussianProcessRegressor() # fit the model model.fit(X, y) # plot the surrogate function plot(X, y, model) |
Running the example first draws the random sample, evaluates it with the noisy objective function, then fits the GP model.
The data sample and a grid of points across the domain evaluated via the surrogate function are then plotted as dots and a line respectively.
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
In this case, as we expected, the plot resembles a crude version of the underlying non-noisy objective function, importantly with a peak around 0.9 where we know the true maxima is located.
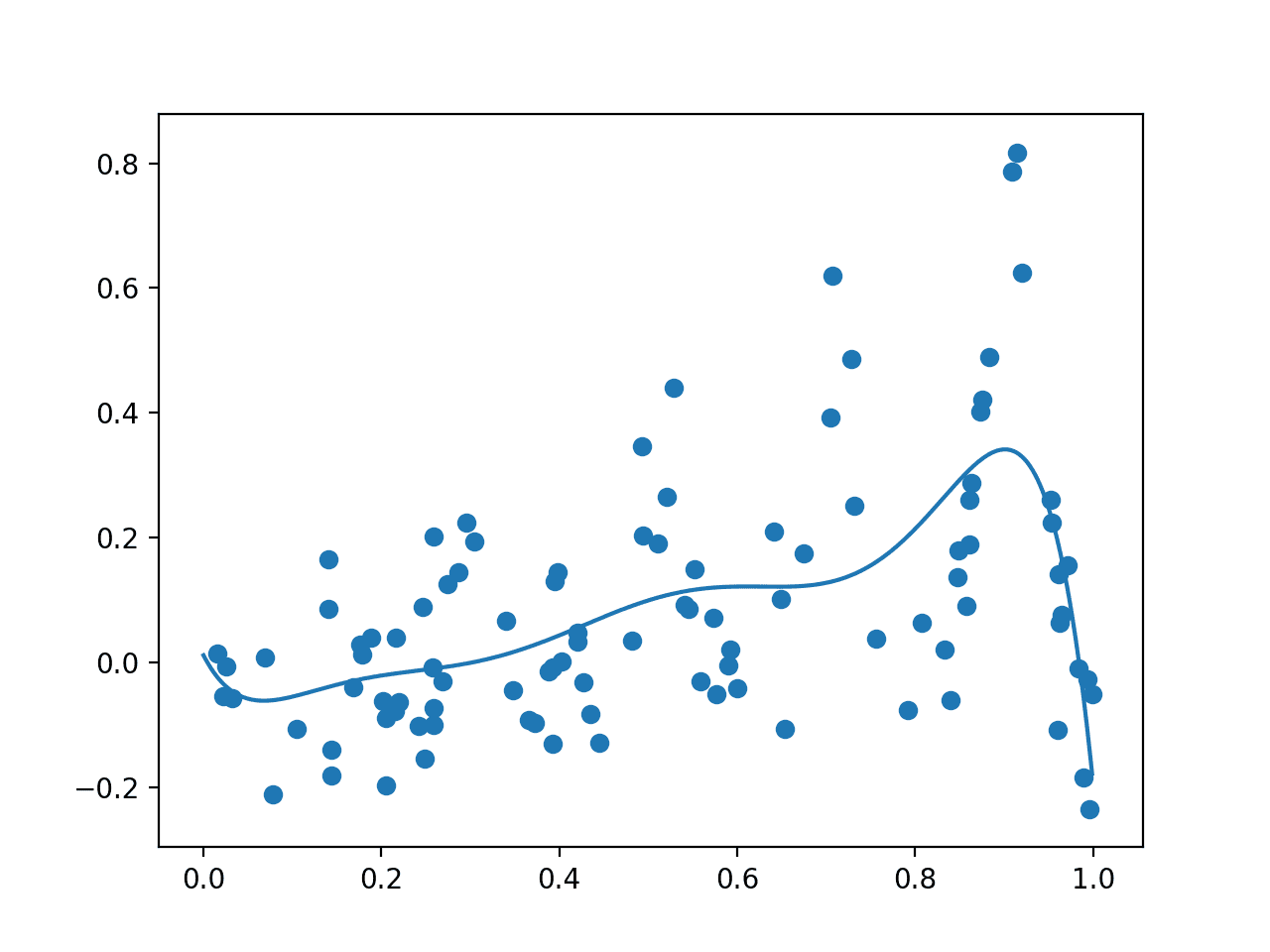
Plot Showing Random Sample With Noisy Evaluation (dots) and Surrogate Function Across the Domain (line).
Next, we must define a strategy for sampling the surrogate function.
Acquisition Function
The surrogate function is used to test a range of candidate samples in the domain.
From these results, one or more candidates can be selected and evaluated with the real, and in normal practice, computationally expensive cost function.
This involves two pieces: the search strategy used to navigate the domain in response to the surrogate function and the acquisition function that is used to interpret and score the response from the surrogate function.
A simple search strategy, such as a random sample or grid-based sample, can be used, although it is more common to use a local search strategy, such as the popular BFGS algorithm. In this case, we will use a random search or random sample of the domain in order to keep the example simple.
This involves first drawing a random sample of candidate samples from the domain, evaluating them with the acquisition function, then maximizing the acquisition function or choosing the candidate sample that gives the best score. The opt_acquisition() function below implements this.
|
1 2 3 4 5 6 7 8 9 10 |
# optimize the acquisition function def opt_acquisition(X, y, model): # random search, generate random samples Xsamples = random(100) Xsamples = Xsamples.reshape(len(Xsamples), 1) # calculate the acquisition function for each sample scores = acquisition(X, Xsamples, model) # locate the index of the largest scores ix = argmax(scores) return Xsamples[ix, 0] |
The acquisition function is responsible for scoring or estimating the likelihood that a given candidate sample (input) is worth evaluating with the real objective function.
We could just use the surrogate score directly. Alternately, given that we have chosen a Gaussian Process model as the surrogate function, we can use the probabilistic information from this model in the acquisition function to calculate the probability that a given sample is worth evaluating.
There are many different types of probabilistic acquisition functions that can be used, each providing a different trade-off for how exploitative (greedy) and explorative they are.
Three common examples include:
- Probability of Improvement (PI).
- Expected Improvement (EI).
- Lower Confidence Bound (LCB).
The Probability of Improvement method is the simplest, whereas the Expected Improvement method is the most commonly used.
In this case, we will use the simpler Probability of Improvement method, which is calculated as the normal cumulative probability of the normalized expected improvement, calculated as follows:
- PI = cdf((mu – best_mu) / stdev)
Where PI is the probability of improvement, cdf() is the normal cumulative distribution function, mu is the mean of the surrogate function for a given sample x, stdev is the standard deviation of the surrogate function for a given sample x, and best_mu is the mean of the surrogate function for the best sample found so far.
We can add a very small number to the standard deviation to avoid a divide by zero error.
The acquisition() function below implements this given the current training dataset of input samples, an array of new candidate samples, and the fit GP model.
|
1 2 3 4 5 6 7 8 9 10 11 |
# probability of improvement acquisition function def acquisition(X, Xsamples, model): # calculate the best surrogate score found so far yhat, _ = surrogate(model, X) best = max(yhat) # calculate mean and stdev via surrogate function mu, std = surrogate(model, Xsamples) mu = mu[:, 0] # calculate the probability of improvement probs = norm.cdf((mu - best) / (std+1E-9)) return probs |
Complete Bayesian Optimization Algorithm
We can tie all of this together into the Bayesian Optimization algorithm.
The main algorithm involves cycles of selecting candidate samples, evaluating them with the objective function, then updating the GP model.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 |
... # perform the optimization process for i in range(100): # select the next point to sample x = opt_acquisition(X, y, model) # sample the point actual = objective(x) # summarize the finding for our own reporting est, _ = surrogate(model, [[x]]) print('>x=%.3f, f()=%3f, actual=%.3f' % (x, est, actual)) # add the data to the dataset X = vstack((X, [[x]])) y = vstack((y, [[actual]])) # update the model model.fit(X, y) |
The complete example is listed below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 |
# example of bayesian optimization for a 1d function from scratch from math import sin from math import pi from numpy import arange from numpy import vstack from numpy import argmax from numpy import asarray from numpy.random import normal from numpy.random import random from scipy.stats import norm from sklearn.gaussian_process import GaussianProcessRegressor from warnings import catch_warnings from warnings import simplefilter from matplotlib import pyplot # objective function def objective(x, noise=0.1): noise = normal(loc=0, scale=noise) return (x**2 * sin(5 * pi * x)**6.0) + noise # surrogate or approximation for the objective function def surrogate(model, X): # catch any warning generated when making a prediction with catch_warnings(): # ignore generated warnings simplefilter("ignore") return model.predict(X, return_std=True) # probability of improvement acquisition function def acquisition(X, Xsamples, model): # calculate the best surrogate score found so far yhat, _ = surrogate(model, X) best = max(yhat) # calculate mean and stdev via surrogate function mu, std = surrogate(model, Xsamples) mu = mu[:, 0] # calculate the probability of improvement probs = norm.cdf((mu - best) / (std+1E-9)) return probs # optimize the acquisition function def opt_acquisition(X, y, model): # random search, generate random samples Xsamples = random(100) Xsamples = Xsamples.reshape(len(Xsamples), 1) # calculate the acquisition function for each sample scores = acquisition(X, Xsamples, model) # locate the index of the largest scores ix = argmax(scores) return Xsamples[ix, 0] # plot real observations vs surrogate function def plot(X, y, model): # scatter plot of inputs and real objective function pyplot.scatter(X, y) # line plot of surrogate function across domain Xsamples = asarray(arange(0, 1, 0.001)) Xsamples = Xsamples.reshape(len(Xsamples), 1) ysamples, _ = surrogate(model, Xsamples) pyplot.plot(Xsamples, ysamples) # show the plot pyplot.show() # sample the domain sparsely with noise X = random(100) y = asarray([objective(x) for x in X]) # reshape into rows and cols X = X.reshape(len(X), 1) y = y.reshape(len(y), 1) # define the model model = GaussianProcessRegressor() # fit the model model.fit(X, y) # plot before hand plot(X, y, model) # perform the optimization process for i in range(100): # select the next point to sample x = opt_acquisition(X, y, model) # sample the point actual = objective(x) # summarize the finding est, _ = surrogate(model, [[x]]) print('>x=%.3f, f()=%3f, actual=%.3f' % (x, est, actual)) # add the data to the dataset X = vstack((X, [[x]])) y = vstack((y, [[actual]])) # update the model model.fit(X, y) # plot all samples and the final surrogate function plot(X, y, model) # best result ix = argmax(y) print('Best Result: x=%.3f, y=%.3f' % (X[ix], y[ix])) |
Running the example first creates an initial random sample of the search space and evaluation of the results. Then a GP model is fit on this data.
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
A plot is created showing the raw observations as dots and the surrogate function across the entire domain. In this case, the initial sample has a good spread across the domain and the surrogate function has a bias towards the part of the domain where we know the optima is located.
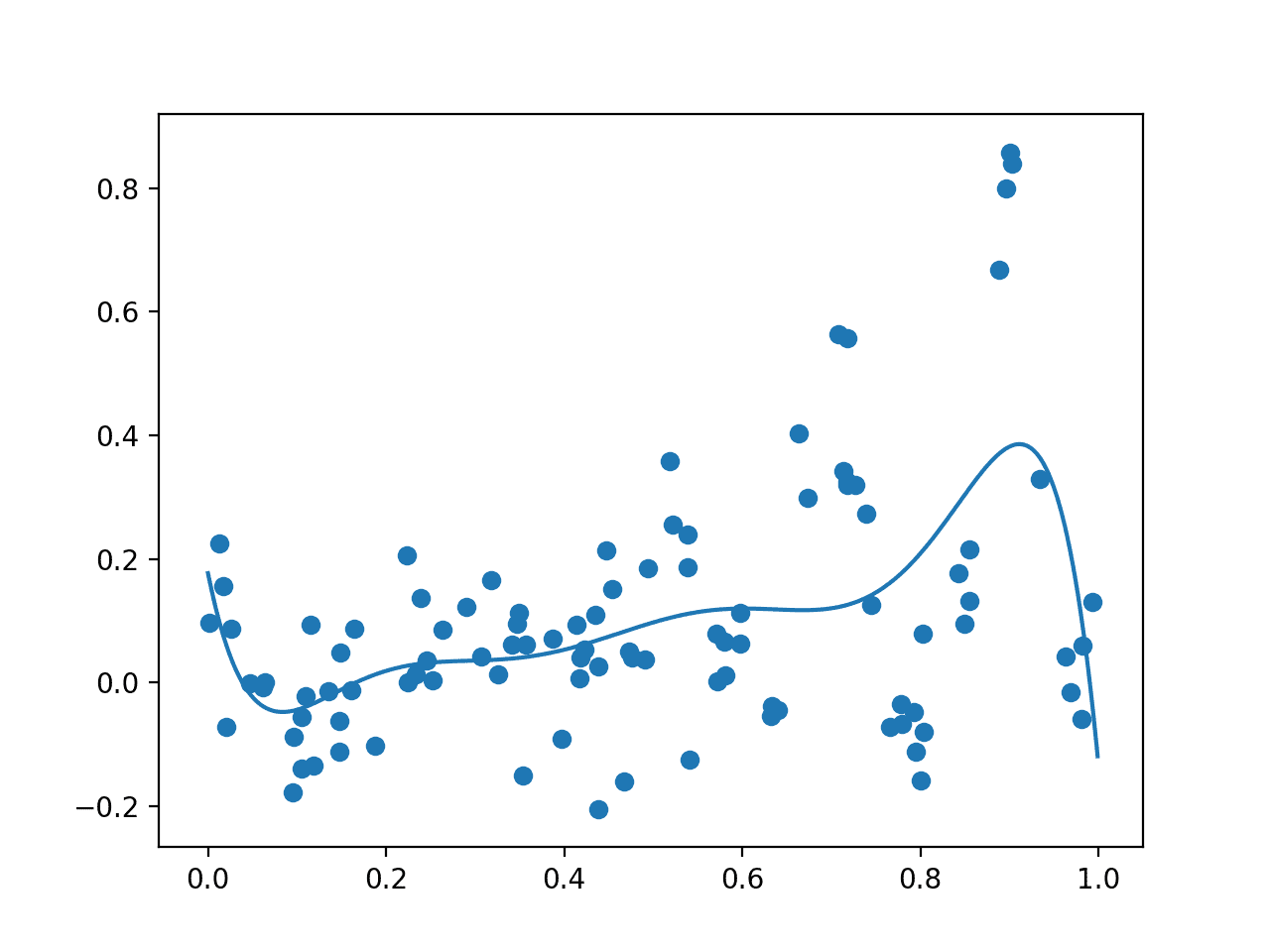
Plot of Initial Sample (dots) and Surrogate Function Across the Domain (line).
The algorithm then iterates for 100 cycles, selecting samples, evaluating them, and adding them to the dataset to update the surrogate function, and over again.
Each cycle reports the selected input value, the estimated score from the surrogate function, and the actual score. Ideally, these scores would get closer and closer as the algorithm converges on one area of the search space.
|
1 2 3 4 5 6 |
... >x=0.922, f()=0.661501, actual=0.682 >x=0.895, f()=0.661668, actual=0.905 >x=0.928, f()=0.648008, actual=0.403 >x=0.908, f()=0.674864, actual=0.750 >x=0.436, f()=0.071377, actual=-0.115 |
Next, a final plot is created with the same form as the prior plot.
This time, all 200 samples evaluated during the optimization task are plotted. We would expect an overabundance of sampling around the known optima, and this is what we see, with may dots around 0.9. We also see that the surrogate function has a stronger representation of the underlying target domain.
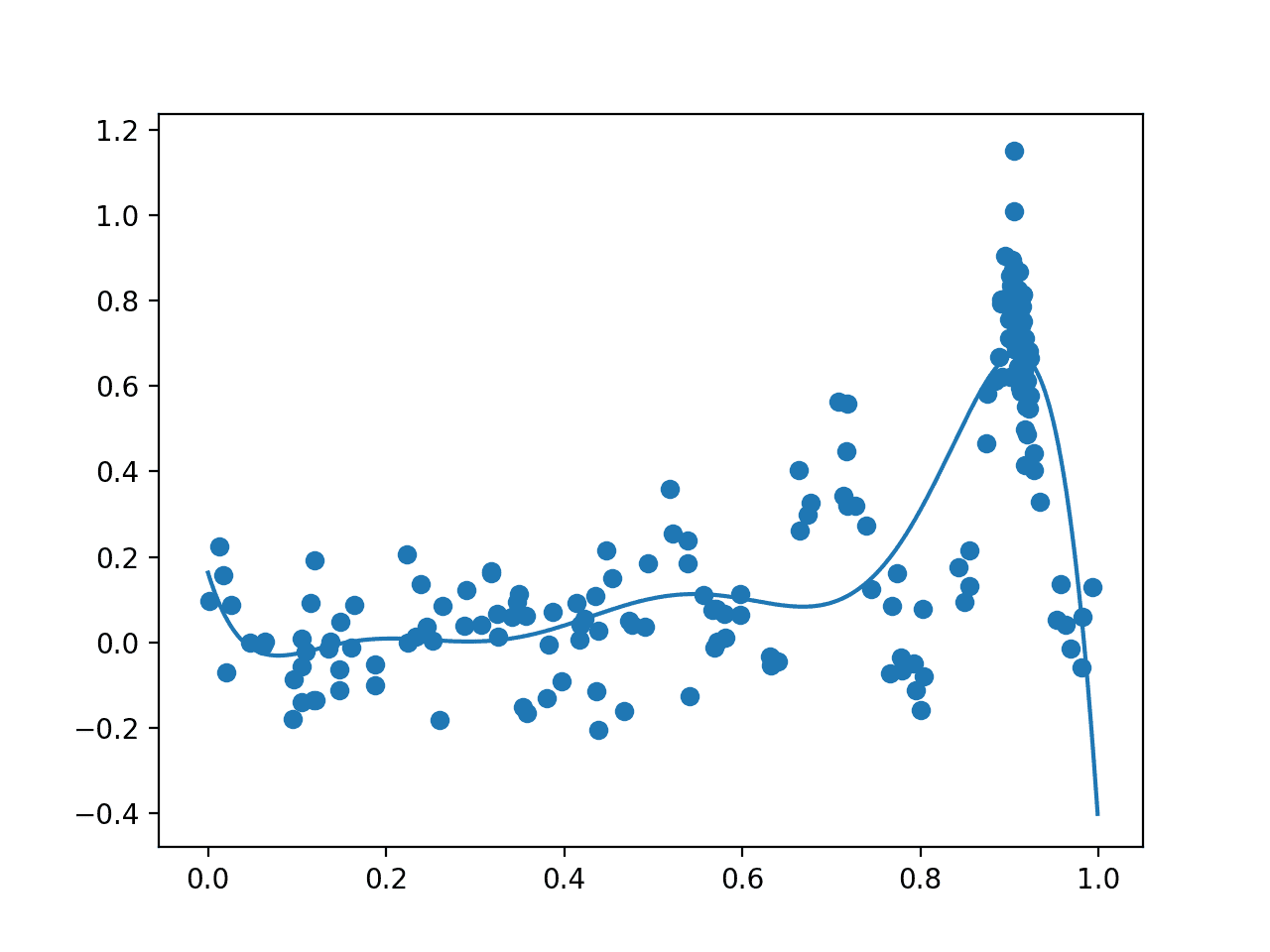
Plot of All Samples (dots) and Surrogate Function Across the Domain (line) after Bayesian Optimization.
Finally, the best input and its objective function score are reported.
We know the optima has an input of 0.9 and an output of 0.810 if there was no sampling noise.
Given the sampling noise, the optimization algorithm gets close in this case, suggesting an input of 0.905.
|
1 |
Best Result: x=0.905, y=1.150 |
Hyperparameter Tuning With Bayesian Optimization
It can be a useful exercise to implement Bayesian Optimization to learn how it works.
In practice, when using Bayesian Optimization on a project, it is a good idea to use a standard implementation provided in an open-source library. This is to both avoid bugs and to leverage a wider range of configuration options and speed improvements.
Two popular libraries for Bayesian Optimization include Scikit-Optimize and HyperOpt. In machine learning, these libraries are often used to tune the hyperparameters of algorithms.
Hyperparameter tuning is a good fit for Bayesian Optimization because the evaluation function is computationally expensive (e.g. training models for each set of hyperparameters) and noisy (e.g. noise in training data and stochastic learning algorithms).
In this section, we will take a brief look at how to use the Scikit-Optimize library to optimize the hyperparameters of a k-nearest neighbor classifier for a simple test classification problem. This will provide a useful template that you can use on your own projects.
The Scikit-Optimize project is designed to provide access to Bayesian Optimization for applications that use SciPy and NumPy, or applications that use scikit-learn machine learning algorithms.
First, the library must be installed, which can be achieved easily using pip; for example:
|
1 |
sudo pip install scikit-optimize |
It is also assumed that you have scikit-learn installed for this example.
Once installed, there are two ways that scikit-optimize can be used to optimize the hyperparameters of a scikit-learn algorithm. The first is to perform the optimization directly on a search space, and the second is to use the BayesSearchCV class, a sibling of the scikit-learn native classes for random and grid searching.
In this example, will use the simpler approach of optimizing the hyperparameters directly.
The first step is to prepare the data and define the model. We will use a simple test classification problem via the make_blobs() function with 500 examples, each with two features and three class labels. We will then use a KNeighborsClassifier algorithm.
|
1 2 3 4 5 |
... # generate 2d classification dataset X, y = make_blobs(n_samples=500, centers=3, n_features=2) # define the model model = KNeighborsClassifier() |
Next, we must define the search space.
In this case, we will tune the number of neighbors (n_neighbors) and the shape of the neighborhood function (p). This requires ranges be defined for a given data type. In this case, they are Integers, defined with the min, max, and the name of the parameter to the scikit-learn model. For your algorithm, you can just as easily optimize Real() and Categorical() data types.
|
1 2 3 |
... # define the space of hyperparameters to search search_space = [Integer(1, 5, name='n_neighbors'), Integer(1, 2, name='p')] |
Next, we need to define a function that will be used to evaluate a given set of hyperparameters. We want to minimize this function, therefore smaller values returned must indicate a better performing model.
We can use the use_named_args() decorator from the scikit-optimize project on the function definition that allows the function to be called directly with a specific set of parameters from the search space.
As such, our custom function will take the hyperparameter values as arguments, which can be provided to the model directly in order to configure it. We can define these arguments generically in python using the **params argument to the function, then pass them to the model via the set_params(**) function.
Now that the model is configured, we can evaluate it. In this case, we will use 5-fold cross-validation on our dataset and evaluate the accuracy for each fold. We can then report the performance of the model as one minus the mean accuracy across these folds. This means that a perfect model with an accuracy of 1.0 will return a value of 0.0 (1.0 – mean accuracy).
This function is defined after we have loaded the dataset and defined the model so that both the dataset and model are in scope and can be used directly.
|
1 2 3 4 5 6 7 8 9 10 |
# define the function used to evaluate a given configuration @use_named_args(search_space) def evaluate_model(**params): # something model.set_params(**params) # calculate 5-fold cross validation result = cross_val_score(model, X, y, cv=5, n_jobs=-1, scoring='accuracy') # calculate the mean of the scores estimate = mean(result) return 1.0 - estimate |
Next, we can perform the optimization.
This is achieved by calling the gp_minimize() function with the name of the objective function and the defined search space.
By default, this function will use a ‘gp_hedge‘ acquisition function that tries to figure out the best strategy, but this can be configured via the acq_func argument. The optimization will also run for 100 iterations by default, but this can be controlled via the n_calls argument.
|
1 2 3 |
... # perform optimization result = gp_minimize(evaluate_model, search_space) |
Once run, we can access the best score via the “fun” property and the best set of hyperparameters via the “x” array property.
|
1 2 3 4 |
... # summarizing finding: print('Best Accuracy: %.3f' % (1.0 - result.fun)) print('Best Parameters: n_neighbors=%d, p=%d' % (result.x[0], result.x[1])) |
Tying this all together, the complete example is listed below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 |
# example of bayesian optimization with scikit-optimize from numpy import mean from sklearn.datasets import make_blobs from sklearn.model_selection import cross_val_score from sklearn.neighbors import KNeighborsClassifier from skopt.space import Integer from skopt.utils import use_named_args from skopt import gp_minimize # generate 2d classification dataset X, y = make_blobs(n_samples=500, centers=3, n_features=2) # define the model model = KNeighborsClassifier() # define the space of hyperparameters to search search_space = [Integer(1, 5, name='n_neighbors'), Integer(1, 2, name='p')] # define the function used to evaluate a given configuration @use_named_args(search_space) def evaluate_model(**params): # something model.set_params(**params) # calculate 5-fold cross validation result = cross_val_score(model, X, y, cv=5, n_jobs=-1, scoring='accuracy') # calculate the mean of the scores estimate = mean(result) return 1.0 - estimate # perform optimization result = gp_minimize(evaluate_model, search_space) # summarizing finding: print('Best Accuracy: %.3f' % (1.0 - result.fun)) print('Best Parameters: n_neighbors=%d, p=%d' % (result.x[0], result.x[1])) |
Running the example executes the hyperparameter tuning using Bayesian Optimization.
The code may report many warning messages, such as:
|
1 |
UserWarning: The objective has been evaluated at this point before. |
This is to be expected and is caused by the same hyperparameter configuration being evaluated more than once.
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
In this case, the model achieved about 97% accuracy via mean 5-fold cross-validation with 3 neighbors and a p-value of 2.
|
1 2 |
Best Accuracy: 0.976 Best Parameters: n_neighbors=3, p=2 |
Further Reading
This section provides more resources on the topic if you are looking to go deeper.
Papers
- A Tutorial on Bayesian Optimization of Expensive Cost Functions, with Application to Active User Modeling and Hierarchical Reinforcement Learning, 2010.
- Practical Bayesian Optimization of Machine Learning Algorithms, 2012.
- A Tutorial on Bayesian Optimization, 2018.
API
- Gaussian Processes, Scikit-Learn API.
- Hyperopt: Distributed Asynchronous Hyper-parameter Optimization
- Scikit-Optimize Project.
- Tuning a scikit-learn estimator with skopt
Articles
- Global optimization, Wikipedia.
- Bayesian optimization, Wikipedia.
- Bayesian optimization, 2018.
- How does Bayesian optimization work?, Quora.
Summary
In this tutorial, you discovered Bayesian Optimization for directed search of complex optimization problems.
Specifically, you learned:
- Global optimization is a challenging problem that involves black box and often non-convex, non-linear, noisy, and computationally expensive objective functions.
- Bayesian Optimization provides a probabilistically principled method for global optimization.
- How to implement Bayesian Optimization from scratch and how to use open-source implementations.
Do you have any questions?
Ask your questions in the comments below and I will do my best to answer.






> # grid-based sample of the domain [0,1]
> X = arange(0, 1, 0.01)
Strictly speaking, this is a grid-based sample of the domain [0, 1), arange works like range in that it doesn’t include the right boundary. It’s minor, but something that has caused me headaches a couple times…
linspace is better for building a grid, IMO.
Great tip, thanks!
Hi
Thanks for another awesome blog post of yours.
I‘m just curious to know about something you may not want to answer about, which would be fine as well.
Do you know already all the details about a given topic you want to write about BEFORE you start writing, or do you first make yourself confident about the topic itself through reading, investigating and trying things out.
Thank you anyway in both cases 🙂
Great question.
Mostly I am familiar with the topics then read/code to write the post. I’ve been doing this for a long time now 🙂
Sometimes I know little and have to spend a few days reading papers/code to get up to speed.
I have some posts outlining how to get up to speed on new topics/algorithms somewhere around here. Ah yes, here under “How to Study/Learn ML Algorithms”
https://machinelearningmastery.com/start-here/#algorithms
Phyton is a high-level programming language , perfect for handling complicated situations.
Bayesian optimization is a non-trivial task, even when applied to simple situations.
The idea of using another programming language , different from Phyton , is a bad idea.
Thanks for sharing.
Hi Jason, thanks for the article. Just want to point out that scikit-optimize is no longer being actively maintained as of Nov 2019 (cf. the project README on github)
Thanks.
The project looks active now and has had many releases this year https://scikit-optimize.github.io/stable/
Agreed, it has picked up!
E.g.:
https://machinelearningmastery.com/scikit-optimize-for-hyperparameter-tuning-in-machine-learning/
Hi Jason
I am new to this but your program doesn’t seem to be able to predict the function at all. With that much data I would have thought it would be enough to predict the second to last peak but it’s completely missing it. I thought maybe the noise is too high for accurate prediction and so I went in and reduced the noise to 0.01 but got the same function. I just don’t see the value here with this example.
Hi Peter, I think you missed the point of the tutorial – or I failed to highlight the point.
Here, we are trying to find the largest peak – it is a optimization problem.
We are not trying to approximate the underlying function, so called function approximation.
One question, you mention that a common acquisition function is the Lower Confidence Bound. However this makes very little sense to me, why would the lower confidence bound be a good metric? it will discourage exploration in places where there is high uncertainty. Maybe you meant Upper Confidence Bound? because that would make a lot of sense as it would do the opposite and encourage exploration where the mean is high and the uncertainty is high.
These are names of methods, perhaps refer to the “further reading” section to discover how they are calculated.
I’m well familiar with both LCB and UCB, and they are computed as mean – sdv and mean + sdv respectively, the lower confidence bound would give you the lower bound of the function given the confidence level, I suppose that the confusion is in the fact that you are maximizing the value (thus you should use UCB), but many paper talk about function minimization, in which case LCB is the appropriate acquisition function to use.
Jason, this is the best post on this topic. Are you doing a covid discount for your learning materials?
Thanks!
Contact me directly if you would like a discount:
https://machinelearningmastery.com/contact/
Hi Jason
Thanks for the amazing article. Can you please add an explanation for Expected Improvement (EI) ? Some of the common HP tuning libraries in python (hyperopt , optuna ) amongst others use EI as the acquisition function…I’ve tried reading a couple of blogs but they are very math heavy and don’t give an intuition about the EI.
what I get so far is that it’s calculating, a expected value ( X x P(x) ) and that EI should be maximized…
can you please explain it in simpler terms?
thanks
You’re welcome.
Great suggestion, thanks!
Hi Jason,
I am not feeling convinced about the part where you calculate the mean vector of model.
Instead of using random sampled data points(surrogate(model, Xsamples)), I think you should use the current training data points.
Why?
Hi,
I am trying to tune model parameters for my ice thickness model. the model has 2 input variables., and 3 model parameters (which i need to tune). I have measured ice thickness data for ~3000 points and want to automate the whole training and testing process. My objective here is to estimate ice thickness with least deviation from the measured ones and find the best set of the three model parameters. So my question is which form of the code should i try ? do i need to havesurrogate and aquisition function here?
Perhaps if there are few parametres to test, you can use a grid search to enumerate them all, rather than use a bayesian search?
Hey Jason, first thank you for your amazing post.
i just had a question. I’m working on Boston house dataset and I’m trying to gain better parameters for ridge ,net and XGBoost using Bayesian optimization. i wonder if the same approach can be implemented on them. i also came across another package called BayesianOptimization . which way do you think is better to approach my problem?
Yes, you can try tuning your model with bayesian optimization.
Hi Jason, can you tell me whether objective function is problem specific (as in the specified function is dependent on the problem we are trying to solve)? If not, does it mean we can specify any function we want because it’s a black box function?
It is not. Correct.
Thank you so much for your great post.
In your example, X, y = make_blobs(n_samples=500, centers=3, n_features=2), if n_features >>2, can this BO still work?
I want to optimize the latent space which dimension is way higher than 2. Your advice is highly appreciated.
Yes. Try it and perhaps compare results to other methods.
Thank you for your reply. I don’t want to search hyperparameters, I want to search for X’s features. That means which X(feature_1, feature_2, …., feature_n) can have maximum y. After optimization, I can get the values of feature_1 to fearure_n instead of model’s hyperparamters.
For example, I use VAE to train tons of molecules. Then I can get a latent space Z. I use this latent space vectors to train a model to predict molecules’ properties, such as molecule’s toxicity. Now I want to search this latent space Z to find a point z with minimum toxicity. Here z is a high dimensional vector.
Sounds great!
But I don’t know how to implement it. Any advice? Thanks.
Perhaps you can start with the example listed above and try adapting it for your needs?
Which part are you stuck on precisely?
Hi Jason,
You provided a really clear explanation.
Could you please explain if Genetic algorithm can be a better one or not when it comes to optimizing the input variables to maximize the objective function?
Also, kindly provide resources to ML techniques that can be used for my problem statement.
Thank you.
Thanks.
It depends on the function being optimized as to what method will work best. Try and compare directly.
What problem statement?
Okay.
Basically I do not have a defined analytical function as y=f(x). I am using a simulator that can provide output y for a given input/input vector x. Thus, the simulator is acting as the function.
The input vector is of 10 variables (x1, x2 etc).
Now, I want to find the optimal values of inputs that maximises the output.
(Bottleneck: It is very costly to obtain large set of data from the simulator. So my idea is to simulate as few points as possible and reach the optimal solution.)
So, I am looking for some method(s) that can solve this problem.
Could you please provide information about what is the best place to start?
Also please enlighten on what are the various optimization techniques and ML techniques that I can explore to try and compare the results?
Yes, you can use the above method and replace the analytical function with the result from your simulator.
I recommend using the library instead of a custom version for speed and reliability.
Okay. Thank you Jason.
Hi Jason,
Thanks for the Post, it was very informative. Perhaps would it be possible to give an explanation of how this Bayesian optimization can be adapted to a classification problem?
Thanks for the suggestion.
Thanks for the nice article.
I guess all the lines by the provided codebase as:
Xsamples = asarray(arange(0, 1, 0.001))
Can be replaced with:
Xsamples = arange(0, 1, 0.001)
Thanks.
Hi Jason, Your posting ahs been really helpful. I have a question ..If probs in aquisition has all zeros then ix = argmax(scores) under opt_aquistion would always take ix=0 .Is this correct?
Thanks.
Yes.
Hi Jason,
Thanks. When will the output of the surrogate model be zero ? ie
In , mu, std = surrogate(model, Xsamples), when will mu be = 0 ? I have an objective function which I have defined as part of surrogate model, made model fit and now predicting for new sample points.. But unable to understand why I am getting zero as predictions ? When will Gaussian Process regressor give a prediction as zero ?
Zero output for the entire domain sounds like a problem. Perhaps there is a bug in your implementation?
Hi PK,
Did you solve the problem? I have the same issue with my surrogate, except that I have an aditional problem, the probs of improvement are always zero too. I implemented the original code of Jason and printed out the probs and it turns out that they are all zeros like in my own model .
Hi Jason,
Thanks for your nice tutorial. it is really helpful.
I have a question, what if I do not have a objective function but some known discrete points, like input [1,2] and output [3] and many other points. if this is my original samples how can I get a objective funtion? Could you give me some advice?
The objective function would be defined with regard to your points – I would expect.
Great tutorial. Finally, someone succeeded to make me understand how BO works! 🙂
I have one minor suggestion.
In this example, we sample X 100 times in [0,1] and add noise with std=0.1. Given the relatively small noise, the maximum of the function will quite often occur at x=0.9. Therefore we already have the maximum in the prior even before we start BO iteration. There’s no problem, but it makes the algorithm less interesting.
What about starting from a subset of X (preferably without the maxima point) as the inial prior?
For example, I did :
X_ans = np.arange(0, 1, 0.001)
y_ans = objective(X_ans)
X = X_ans[::71] # <— sub sample to avoid the maximum
y = y_ans[::71]
X = X.reshape(-1,1)
y = y.reshape(-1,1)
… and so on…
Now, my BO algorithm can get much closer to the 'real' maximum than the initial data I had, and I found it more interesting.
+ coloring new points made me feel even better!
Again, thanks a lot for the great tutorial.
Thanks.
Great suggestion.
Hi Jason! Thank you for this post, helped me to understand better the algorithm!
I have a similar question that Xuan Wang posted. I don’t have a explicit analytical objetive function, I actually used a Gaussian Process Regressor to model my data and thus assume that this is my true objetive function. So, what I’m trying to find is the maximum value of my gaussian process model.
In this case, should I use both my objetive function and the model of the Bayesian Optimization as the same Gaussian process regressor (with the same parameters)? I obtained good results by using this, but I’m not 100% confident that this is correct.
You’re welcome.
Not sure I follow, you must have an evaluation of something for the Gaussian Process Regressor to be approximating.
I have 4D data (x1, x2, x3 and y), which are readings from sensors in field, and I used a Gaussian Process Regressor to derive a black box model such that y = GPR(x1,x2,x3).
Now, my final goal is to find the maximum value of GRP(x1,x2,x3), for example, assuming the only way to evaluate y is from the gaussian process. Bayesian optimization still applies in this case? The algorithm would have two gaussian processes, adding the one which is used for the maximization.
Thanks for the fast reply!
Perhaps try it and see.
Hi Jason,
Thanks for one more quite enlightening article.
I have a question regarding the evaluation of the objective function. You obtain the ‘actual’ optimum from the noisy version at each cycle. Why?
I thought first that you would minimize the analytical model. It is not seem to be the case. Are you instead generating ‘synthetic data’ and trying to optimize some ‘unknown decision function’?
If so, how would I perform this in a real scenario? I mean, supposing I do not know the analytical form of the objective function (just have the data points). How should I evaluate the “actual” candidate at each cycle provided that I just have the data?
This problem setup assumes a noisy function evaluation. If this is not the case for your problem, bayes opt might not be appropriate.
Hello,
Thank you for the article.
I have a question : How can I define my search space if my parameter to optimize is not an integer but a tuple of integers ? i.e. ARIMA’s order = (p,d,q)
I’d like to perform optimization on this tridimensional space but I cannot define my search space properly.
It would be all combinations, e.g. nested for loops, for example:
https://machinelearningmastery.com/grid-search-arima-hyperparameters-with-python/
I just read the article you sent.
However, I feel like having nested for loops is too resource-consuming versus implementing a search algorithm.
If I want to tune a SARIMAX (p,d,q,P,D,Q,m) model and make 7 nested for loops, my computation time will increase in o(x^7).
Isn’t there a way to apply stochastic search methods to ARIMA tuning ?
Not really, see this example:
https://machinelearningmastery.com/how-to-grid-search-sarima-model-hyperparameters-for-time-series-forecasting-in-python/
Thank you very much for the quick answer ! I’ve read avidly a lot of your articles, and they are truly a gold mine.
You’re welcome!
Hello Jason,
Thank you for the great.
We have an use case where a black box optimization provides solution to the problem(finding the parameters of physics based model whose mathematical properties are unknown ).
Currently I don’t have knowledge about internal working of Bayesian optimization. I will study and try to implement. This post will be a great help to start the implementation.
I have one question. Does Bayesian optimization needs lots of data like supervised learning? according my naive understanding, data is not required for Bayesian optimization. Algorithm itself samples and searches optimum parameters. What it needs is just an evaluation of the output(parameters).
Let me know if my understanding is correct.
thank you
Thank You for the great post*
You’re welcome.
It may or may not require many samples of the problem, it really depends on the problem.
Thank You for the quick reply. Require samples here mean algorithm itself samples? Or do we have to provide prior distribution.(May be my question doesn’t make sense as I don’t know the working of the algorithm)
Thank you
Sorry “samples” means evaluations of inputs against your objective function.
Thank You for your reply. In this implementation, Objective function is continuous.
Is it required that it has be continuous? in my problem which I am solving(finding best low pass filer co-efficient) objective function is discrete in nature. It is not possible determine objective function value for intermediate values. in this case how do we formulate objective function?
Yes, I believe so for this implementation. Strictly, I don’t think so.
Hey Jason
Great blog and I have just downloaded the full set of book.
I come from the decision under uncertainty space – many years ago – and used the Hooke-Jeeves method before, but it performs very poorly with uncertainty/variability. So I am very keen to try out the Bayesian Optimization.
For me a single variable example is quite limited. As I am quite new to Python, I was wondering if you could extend your example to the Rosenbrock Banana Function, which uses 2 variables. https://en.wikipedia.org/wiki/Rosenbrock_function
Thank you for your suggestion. Indeed, it should not be difficult to extend the example. If you can spend some time on an introductory book in Python, that should help you further in learning.
Thank you for sharing. I would like to point out that the GP is not tuned in the sense where its hyperparameters are not estimated as iterations. This is because you use the following kernel for the GP :
ConstantKernel(1.0, constant_value_bounds=”fixed” * RBF(1.0, length_scale_bounds=”fixed”) which is the default kernel.
Another choice that works better is : kernel = C(1.0, (1e-3, 1e3)) * RBF(10, (1e-2, 1e2)), then write model = GaussianProcessRegressor(kernel=kernel).
Thanks for pointing out!
Hello Dambling. Could you please elaborate more on that?
Hi,
Thanks a lot for e detailed explanation. It would be really helpful if you can guide me to resources that compare and contrast Bayesian Optimization and Reinforcement Learning. Especially when I want to understand a sequence of inputs leading to an optima, which would be a better approach : RL or SMBO ? Also , I am curious in knowing the specific use cases of Bayesian optimization other than hyperparameter tuning.
Thanks a lot !!
Hi Yashaswini…You may want to review the following:
https://towardsdatascience.com/reinforcement-learning-vs-bayesian-optimization-when-to-use-what-be32fd6e83da
Regards,
Lol,
> model = GaussianProcessRegressor()
> model.fit()
I really appreciate you posted this article, but this is not “Bayesian Optimization from Scratch”. Maybe if scikit-learn is implemented in Scratch? But I kind of doubt it.
Thank you for your feedback, Matthew!
Hello, James Carmichael!
Fantastic article!
At “Acquisition Function” section, I can’t understand the role of cdf() function.
> # Raw code
> probs = cdf((mu – best) / (std+1E-9))
>
> # opt_acquisition() find probs greatest probs point.
> # If the x is greatest, the cdf value also is greatest.
> # cdf([0.1, 0.1, 0.4, 0.5, 0.2, 0.5, 0.7, 0.1, 0.1])
> # array([0.53982784, 0.53982784, 0.65542174, 0.69146246, 0.57925971,
> # 0.69146246, 0.75803635, 0.53982784, 0.53982784])
> # I wonder if I could write it this way and get the same result
> probs = (mu – best) / (std+1E-9)
Hi Song…Please see the following that covers the Cumulative Distribution Function:
https://machinelearningmastery.com/empirical-distribution-function-in-python/#:~:text=The%20CDF%20returns%20the%20expected,Kernel%20Density%20Estimation%20(KDE).
Hello, James Carmichael !
The role of CDF I have seen this article of yours before and it was very useful for me. I guess I didn’t express my point clearly. I mean is that I can’t understand the cdf() function in this article’s “acquisition” function.
The raw code in “acquisition” function is:
> probs = norm.cdf( (mu – best) / (std+1E-9) )
If I only delete the cdf() function, the code will give same result.
> probs = (mu – best) / (std+1E-9)
Because the “opt_acquisition” function get the “acquisition” function’s return vector and calculate the greatest value index. However, the cdf function give the value’s cumulative. If the value is greatest and the return value of cdf() is also greatest, so delete the cdf maybe get the same result.
> # This is a cdf test
> from scipy.stats import norm
> x = [0.1, 0.1, 0.4, 0.5,
0.2, 0.5, 0.7, 0.1,
0.1]
> norm.cdf( x )
# array([0.53982784, 0.53982784, 0.65542174, 0.69146246,
# 0.57925971, 0.69146246, 0.75803635, 0.53982784,
# 0.53982784 ]
Thanks!
Dear Jason Brownlee,
Thank you for your excellent tutorial, I really like the hands-on approaches to make the ideas 100% clear. I have one question though about your example, more explicitly about the acquisition-function, which I copied from your post to below:
# probability of improvement acquisition function
def acquisition(X, Xsamples, model):
# calculate the best surrogate score found so far
yhat, _ = surrogate(model, X)
best = max(yhat)
# calculate mean and stdev via surrogate function
mu, std = surrogate(model, Xsamples)
mu = mu[:, 0]
# calculate the probability of improvement
probs = norm.cdf((mu – best) / (std+1E-9))
return probs
I’m having a bit trouble understanding what is going on here, that is the selection logic (the criteria by which to select the next point) but I try to break it down line by line:
1. In “yhat, _ = surrogate(model, X)”, we calculate the model fitted means (i.e. the predictions at observed x-locations) of our observed (X,y) data.
2. The “best = max(yhat)” corresponds to the maximum mean prediction.
3. At “mu, std = surrogate(model, Xsamples)” we calculate the predicted means and stds at the new x-locations (Xsamples) from which we wish to select the optimal new sample location.
4. At “probs = norm.cdf((mu – best) / (std+1E-9))” we calculate the CDF probability values, which is maximized when the standardized distance of “mu” to “best”, that is the z-score, is as positive as possible. Let “mu_best” be the one in “mu” with this property.
Now where I get lost is in perhaps steps 2 and 4. I don’t get this selection logic, why would we want to sample next at position x, which has the mean prediction of “mu_best”?
Lets assume the predictive stds are equal. In this scenario, the PI-rule guides us select to sample from x-location where the “mu” is as large as possible (e.g. mu >> best). So we always select sample points with as large predicted y-values as possible.
Why would this be a good idea? This what I don’t understand. Thank you in advance.
P.S. just to add to my previous question, I don’t understand in what sense the “Probability of Improvement”-rule is improving our selections, in what way can we improve our estimated surrogate model by always selecting e.g. the x-locations with largest predicted means?
Hello Jjep…The following may help further clarify:
https://towardsdatascience.com/a-conceptual-explanation-of-bayesian-model-based-hyperparameter-optimization-for-machine-learning-b8172278050f
Dear James Carmichael,
Thank you for your reply and the link. I found a good explanation from other source:
https://www.borealisai.com/en/blog/tutorial-8-bayesian-optimization/
“The probability of improvement considers the best function value f(x) that we have observed so far (horizontal dashed line) and measures the probability that a sample at the new point will exceed this value.”
So, the terms “best” and “improvement” to my opinion might be a bit misleading here. “Best” is actually the maximum value of the current fitted model, but I don’t see how it is the “best” in some sense. Also the “improvement” I think should be more clear. I don’t understand how the rule: “sample at the new point, which will exceed best f(x) most probably” is considered improvement.
P.S.
adding to previous: I think the “best” and “improvement”-terms would then make sense if the function f would be some kind of a score / performance function, what you think?
One note, the line y = np.vstack((y, [[actual]])) is incorrect. We do not know the actual value derived from the objective function. it should be y = np.vstack((y, [[est]]).
Mistake on previous comment. appending the estimate means you never update the surrogate function. Appending the actual means you have ground truth. What should be appended is the actual value + noise.
Thank you for the feedback Tim!
Thank you for the great explanation!
I am confused when you say acquisition function is the likelihood.
Can you map the relationship between each component(prior, likelihood, posterior) in Bayes Therom and the concepts(surrogate model, acquisition function) you introduced here?
Isn’t our goal trying to maximize the posterior probability? then that means acquisition function corresponds to posterior? but Idk how this is determined, and if so, what is the prior here
p(H|E) = p(E|H)*p(H) = p(E|H)*surrogate(H) = acquisition???
Hi Moon…the following resource will hopefully add clarity:
https://arxiv.org/abs/1807.02811
Hello,
Thanks for the great tutorial. Do you know about another library that I can use to call my own surrogate model (let say, I want to use a DNN)?
Hi Hanser…You may find the following resources of interest:
https://towardsdatascience.com/bayesian-hyperparameter-optimization-for-a-deep-neural-network-590be92bb0c0
https://ieeexplore.ieee.org/document/9612061
Hi James,
Thanks for the resources. The options presented in these post try to tune the hyperparams of a DNN using Gaussian process as the surrogate. However, what I want to do is to use a DNN as a surrogate model (or any other surrogate that I can build myself).
Hi James,
Thanks again for this great tutorial. I have implemented the original script of the post (the “how to perform Bayes Optimization” section) and I found out that the probabilities of improvement (probs in the code) are always zero. At first, I supposed that this happens because the optima is already present in the initial dataset, and therefore all the other samples are not better points. However, I reduced significantly the number of intial samples (just 10 or 20 instead of 100) to avoid catching the optima and have optimization space, but the probs keep being zero. Do you know why that?
Thanks for a great tutorial and a fantastic site.
I have a two questions:
– Is there a principled way to think about the size of the noise parameter?
– Could the sample process in opt_aquisition be improved to better catch prior belief?
Also, it would be nice with an ebook on TensorFlow Probabilty! 🙂 Keep up the fantastic work!
Hi,
I have one question regarding objective function in gp_minimize.
gp_minimize expects for fitness function scalar. But how to use gp_minimize when we have multi output neural network (for one or more input we forcast multi output)? In that case, in order to tune hyperparameters we will have multiple objective (fitness) functions.
Thank you!
Hi Jane…The following discussion provides some interesting ideas in this area:
https://stats.stackexchange.com/questions/309287/multivariate-gaussian-processing-bayesian-optimization-with-multiple-features
Thank you very much for your fast response. I researched a bit and found out that there are several libraries for multi-objective Bayesian optimization using the Gaussian process. Do you have a recommendation on what library is currently most suitable for this?
Thank you!
Hi Jane…You are very welcome! There are many great options such as the following:
https://github.com/bayesian-optimization/BayesianOptimization
hi Jason, I have a query regarding bayesian optimization, can I use it to find the maximum value of output and corresponding feature (input) values predicted by Machine learning algorithms such as multiple linear regression, random forest etc…
Hi Usama…Yes, Bayesian optimization can be used to find the maximum value of the output and the corresponding feature (input) values predicted by machine learning algorithms such as multiple linear regression, random forest, and others. This optimization method is particularly useful for optimizing complex functions that are expensive to evaluate, and it’s well-suited for hyperparameter tuning in machine learning models.
Bayesian optimization works by building a probabilistic model of the function you’re trying to optimize and uses that model to make intelligent guesses about where the optimal points might lie. The key idea is to use the probabilistic model to balance the exploration of unvisited areas of the search space (where the function might attain higher values) against the exploitation of known areas (where the function has already been observed to attain high values).
In the context of machine learning models like multiple linear regression, random forest, etc., Bayesian optimization can be employed to automatically find the set of hyperparameters (or input features for feature engineering tasks) that maximizes the performance of the model. This is done by considering the performance of the model (e.g., accuracy, F1 score, etc.) as the function to be optimized.
For example, if you’re using a random forest model, Bayesian optimization can help you find the optimal number of trees, depth of trees, minimum samples per leaf, etc., that maximize your model’s predictive accuracy. Similarly, for multiple linear regression, it might help in selecting features or tuning other parameters that influence model performance.
To use Bayesian optimization for these purposes, you would:
1. **Define the objective function**: This is the function that takes a set of parameters as input and returns a performance metric (e.g., accuracy) as output, which you want to maximize.
2. **Choose a domain space**: This is the range of values over which you want to optimize the parameters.
3. **Specify a surrogate model**: This is the probabilistic model that approximates the objective function. Gaussian Processes are commonly used, but there are other choices too.
4. **Decide on an acquisition function**: This function is used to decide where to sample next, balancing exploration and exploitation.
Bayesian optimization then iteratively selects the next set of parameters to evaluate by optimizing the acquisition function, updates the surrogate model with the results, and repeats the process until a stopping criterion is met.
It’s a powerful tool, especially when the evaluations of the objective function are very costly (in terms of time or resources), and when the parameter space is high-dimensional and difficult to navigate with simpler optimization methods.