Logistic regression is the go-to linear classification algorithm for two-class problems.
It is easy to implement, easy to understand and gets great results on a wide variety of problems, even when the expectations the method has of your data are violated.
In this tutorial, you will discover how to implement logistic regression with stochastic gradient descent from scratch with Python.
After completing this tutorial, you will know:
- How to make predictions with a logistic regression model.
- How to estimate coefficients using stochastic gradient descent.
- How to apply logistic regression to a real prediction problem.
Kick-start your project with my new book Machine Learning Algorithms From Scratch, including step-by-step tutorials and the Python source code files for all examples.
Let’s get started.
- Update Jan/2017: Changed the calculation of fold_size in cross_validation_split() to always be an integer. Fixes issues with Python 3.
- Update Mar/2018: Added alternate link to download the dataset as the original appears to have been taken down.
- Update Aug/2018: Tested and updated to work with Python 3.6.

How To Implement Logistic Regression With Stochastic Gradient Descent From Scratch With Python
Photo by Ian Sane, some rights reserved.
Description
This section will give a brief description of the logistic regression technique, stochastic gradient descent and the Pima Indians diabetes dataset we will use in this tutorial.
Logistic Regression
Logistic regression is named for the function used at the core of the method, the logistic function.
Logistic regression uses an equation as the representation, very much like linear regression. Input values (X) are combined linearly using weights or coefficient values to predict an output value (y).
A key difference from linear regression is that the output value being modeled is a binary value (0 or 1) rather than a numeric value.
|
1 |
yhat = e^(b0 + b1 * x1) / (1 + e^(b0 + b1 * x1)) |
This can be simplified as:
|
1 |
yhat = 1.0 / (1.0 + e^(-(b0 + b1 * x1))) |
Where e is the base of the natural logarithms (Euler’s number), yhat is the predicted output, b0 is the bias or intercept term and b1 is the coefficient for the single input value (x1).
The yhat prediction is a real value between 0 and 1, that needs to be rounded to an integer value and mapped to a predicted class value.
Each column in your input data has an associated b coefficient (a constant real value) that must be learned from your training data. The actual representation of the model that you would store in memory or in a file are the coefficients in the equation (the beta value or b’s).
The coefficients of the logistic regression algorithm must be estimated from your training data.
Stochastic Gradient Descent
Gradient Descent is the process of minimizing a function by following the gradients of the cost function.
This involves knowing the form of the cost as well as the derivative so that from a given point you know the gradient and can move in that direction, e.g. downhill towards the minimum value.
In machine learning, we can use a technique that evaluates and updates the coefficients every iteration called stochastic gradient descent to minimize the error of a model on our training data.
The way this optimization algorithm works is that each training instance is shown to the model one at a time. The model makes a prediction for a training instance, the error is calculated and the model is updated in order to reduce the error for the next prediction.
This procedure can be used to find the set of coefficients in a model that result in the smallest error for the model on the training data. Each iteration, the coefficients (b) in machine learning language are updated using the equation:
|
1 |
b = b + learning_rate * (y - yhat) * yhat * (1 - yhat) * x |
Where b is the coefficient or weight being optimized, learning_rate is a learning rate that you must configure (e.g. 0.01), (y – yhat) is the prediction error for the model on the training data attributed to the weight, yhat is the prediction made by the coefficients and x is the input value.
Pima Indians Diabetes Dataset
The Pima Indians dataset involves predicting the onset of diabetes within 5 years in Pima Indians given basic medical details.
It is a binary classification problem, where the prediction is either 0 (no diabetes) or 1 (diabetes).
It contains 768 rows and 9 columns. All of the values in the file are numeric, specifically floating point values. Below is a small sample of the first few rows of the problem.
|
1 2 3 4 5 6 |
6,148,72,35,0,33.6,0.627,50,1 1,85,66,29,0,26.6,0.351,31,0 8,183,64,0,0,23.3,0.672,32,1 1,89,66,23,94,28.1,0.167,21,0 0,137,40,35,168,43.1,2.288,33,1 ... |
Predicting the majority class (Zero Rule Algorithm), the baseline performance on this problem is 65.098% classification accuracy.
Download the dataset and save it to your current working directory with the filename pima-indians-diabetes.csv.
Tutorial
This tutorial is broken down into 3 parts.
- Making Predictions.
- Estimating Coefficients.
- Diabetes Prediction.
This will provide the foundation you need to implement and apply logistic regression with stochastic gradient descent on your own predictive modeling problems.
1. Making Predictions
The first step is to develop a function that can make predictions.
This will be needed both in the evaluation of candidate coefficient values in stochastic gradient descent and after the model is finalized and we wish to start making predictions on test data or new data.
Below is a function named predict() that predicts an output value for a row given a set of coefficients.
The first coefficient in is always the intercept, also called the bias or b0 as it is standalone and not responsible for a specific input value.
|
1 2 3 4 5 6 |
# Make a prediction with coefficients def predict(row, coefficients): yhat = coefficients[0] for i in range(len(row)-1): yhat += coefficients[i + 1] * row[i] return 1.0 / (1.0 + exp(-yhat)) |
We can contrive a small dataset to test our predict() function.
|
1 2 3 4 5 6 7 8 9 10 11 |
X1 X2 Y 2.7810836 2.550537003 0 1.465489372 2.362125076 0 3.396561688 4.400293529 0 1.38807019 1.850220317 0 3.06407232 3.005305973 0 7.627531214 2.759262235 1 5.332441248 2.088626775 1 6.922596716 1.77106367 1 8.675418651 -0.242068655 1 7.673756466 3.508563011 1 |
Below is a plot of the dataset using different colors to show the different classes for each point.
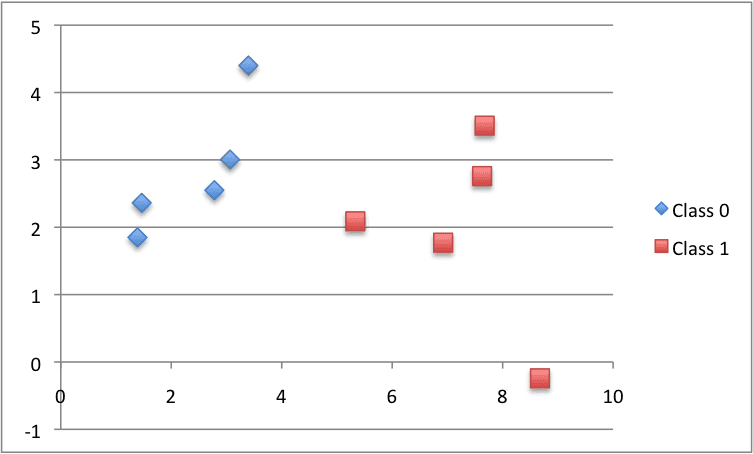
Small Contrived Classification Dataset
We can also use previously prepared coefficients to make predictions for this dataset.
Putting this all together we can test our predict() function below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 |
# Make a prediction from math import exp # Make a prediction with coefficients def predict(row, coefficients): yhat = coefficients[0] for i in range(len(row)-1): yhat += coefficients[i + 1] * row[i] return 1.0 / (1.0 + exp(-yhat)) # test predictions dataset = [[2.7810836,2.550537003,0], [1.465489372,2.362125076,0], [3.396561688,4.400293529,0], [1.38807019,1.850220317,0], [3.06407232,3.005305973,0], [7.627531214,2.759262235,1], [5.332441248,2.088626775,1], [6.922596716,1.77106367,1], [8.675418651,-0.242068655,1], [7.673756466,3.508563011,1]] coef = [-0.406605464, 0.852573316, -1.104746259] for row in dataset: yhat = predict(row, coef) print("Expected=%.3f, Predicted=%.3f [%d]" % (row[-1], yhat, round(yhat))) |
There are two inputs values (X1 and X2) and three coefficient values (b0, b1 and b2). The prediction equation we have modeled for this problem is:
|
1 |
y = 1.0 / (1.0 + e^(-(b0 + b1 * X1 + b2 * X2))) |
or, with the specific coefficient values we chose by hand as:
|
1 |
y = 1.0 / (1.0 + e^(-(-0.406605464 + 0.852573316 * X1 + -1.104746259 * X2))) |
Running this function we get predictions that are reasonably close to the expected output (y) values and when rounded make correct predictions of the class.
|
1 2 3 4 5 6 7 8 9 10 |
Expected=0.000, Predicted=0.299 [0] Expected=0.000, Predicted=0.146 [0] Expected=0.000, Predicted=0.085 [0] Expected=0.000, Predicted=0.220 [0] Expected=0.000, Predicted=0.247 [0] Expected=1.000, Predicted=0.955 [1] Expected=1.000, Predicted=0.862 [1] Expected=1.000, Predicted=0.972 [1] Expected=1.000, Predicted=0.999 [1] Expected=1.000, Predicted=0.905 [1] |
Now we are ready to implement stochastic gradient descent to optimize our coefficient values.
2. Estimating Coefficients
We can estimate the coefficient values for our training data using stochastic gradient descent.
Stochastic gradient descent requires two parameters:
- Learning Rate: Used to limit the amount each coefficient is corrected each time it is updated.
- Epochs: The number of times to run through the training data while updating the coefficients.
These, along with the training data will be the arguments to the function.
There are 3 loops we need to perform in the function:
- Loop over each epoch.
- Loop over each row in the training data for an epoch.
- Loop over each coefficient and update it for a row in an epoch.
As you can see, we update each coefficient for each row in the training data, each epoch.
Coefficients are updated based on the error the model made. The error is calculated as the difference between the expected output value and the prediction made with the candidate coefficients.
There is one coefficient to weight each input attribute, and these are updated in a consistent way, for example:
|
1 |
b1(t+1) = b1(t) + learning_rate * (y(t) - yhat(t)) * yhat(t) * (1 - yhat(t)) * x1(t) |
The special coefficient at the beginning of the list, also called the intercept, is updated in a similar way, except without an input as it is not associated with a specific input value:
|
1 |
b0(t+1) = b0(t) + learning_rate * (y(t) - yhat(t)) * yhat(t) * (1 - yhat(t)) |
Now we can put all of this together. Below is a function named coefficients_sgd() that calculates coefficient values for a training dataset using stochastic gradient descent.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 |
# Estimate logistic regression coefficients using stochastic gradient descent def coefficients_sgd(train, l_rate, n_epoch): coef = [0.0 for i in range(len(train[0]))] for epoch in range(n_epoch): sum_error = 0 for row in train: yhat = predict(row, coef) error = row[-1] - yhat sum_error += error**2 coef[0] = coef[0] + l_rate * error * yhat * (1.0 - yhat) for i in range(len(row)-1): coef[i + 1] = coef[i + 1] + l_rate * error * yhat * (1.0 - yhat) * row[i] print('>epoch=%d, lrate=%.3f, error=%.3f' % (epoch, l_rate, sum_error)) return coef |
You can see, that in addition, we keep track of the sum of the squared error (a positive value) each epoch so that we can print out a nice message each outer loop.
We can test this function on the same small contrived dataset from above.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 |
from math import exp # Make a prediction with coefficients def predict(row, coefficients): yhat = coefficients[0] for i in range(len(row)-1): yhat += coefficients[i + 1] * row[i] return 1.0 / (1.0 + exp(-yhat)) # Estimate logistic regression coefficients using stochastic gradient descent def coefficients_sgd(train, l_rate, n_epoch): coef = [0.0 for i in range(len(train[0]))] for epoch in range(n_epoch): sum_error = 0 for row in train: yhat = predict(row, coef) error = row[-1] - yhat sum_error += error**2 coef[0] = coef[0] + l_rate * error * yhat * (1.0 - yhat) for i in range(len(row)-1): coef[i + 1] = coef[i + 1] + l_rate * error * yhat * (1.0 - yhat) * row[i] print('>epoch=%d, lrate=%.3f, error=%.3f' % (epoch, l_rate, sum_error)) return coef # Calculate coefficients dataset = [[2.7810836,2.550537003,0], [1.465489372,2.362125076,0], [3.396561688,4.400293529,0], [1.38807019,1.850220317,0], [3.06407232,3.005305973,0], [7.627531214,2.759262235,1], [5.332441248,2.088626775,1], [6.922596716,1.77106367,1], [8.675418651,-0.242068655,1], [7.673756466,3.508563011,1]] l_rate = 0.3 n_epoch = 100 coef = coefficients_sgd(dataset, l_rate, n_epoch) print(coef) |
We use a larger learning rate of 0.3 and train the model for 100 epochs, or 100 exposures of the coefficients to the entire training dataset.
Running the example prints a message each epoch with the sum squared error for that epoch and the final set of coefficients.
|
1 2 3 4 5 6 |
>epoch=95, lrate=0.300, error=0.023 >epoch=96, lrate=0.300, error=0.023 >epoch=97, lrate=0.300, error=0.023 >epoch=98, lrate=0.300, error=0.023 >epoch=99, lrate=0.300, error=0.022 [-0.8596443546618897, 1.5223825112460005, -2.218700210565016] |
You can see how error continues to drop even in the final epoch. We could probably train for a lot longer (more epochs) or increase the amount we update the coefficients each epoch (higher learning rate).
Experiment and see what you come up with.
Now, let’s apply this algorithm on a real dataset.
3. Diabetes Prediction
In this section, we will train a logistic regression model using stochastic gradient descent on the diabetes dataset.
The example assumes that a CSV copy of the dataset is in the current working directory with the filename pima-indians-diabetes.csv.
The dataset is first loaded, the string values converted to numeric and each column is normalized to values in the range of 0 to 1. This is achieved with the helper functions load_csv() and str_column_to_float() to load and prepare the dataset and dataset_minmax() and normalize_dataset() to normalize it.
We will use k-fold cross validation to estimate the performance of the learned model on unseen data. This means that we will construct and evaluate k models and estimate the performance as the mean model performance. Classification accuracy will be used to evaluate each model. These behaviors are provided in the cross_validation_split(), accuracy_metric() and evaluate_algorithm() helper functions.
We will use the predict(), coefficients_sgd() functions created above and a new logistic_regression() function to train the model.
Below is the complete example.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 |
# Logistic Regression on Diabetes Dataset from random import seed from random import randrange from csv import reader from math import exp # Load a CSV file def load_csv(filename): dataset = list() with open(filename, 'r') as file: csv_reader = reader(file) for row in csv_reader: if not row: continue dataset.append(row) return dataset # Convert string column to float def str_column_to_float(dataset, column): for row in dataset: row[column] = float(row[column].strip()) # Find the min and max values for each column def dataset_minmax(dataset): minmax = list() for i in range(len(dataset[0])): col_values = [row[i] for row in dataset] value_min = min(col_values) value_max = max(col_values) minmax.append([value_min, value_max]) return minmax # Rescale dataset columns to the range 0-1 def normalize_dataset(dataset, minmax): for row in dataset: for i in range(len(row)): row[i] = (row[i] - minmax[i][0]) / (minmax[i][1] - minmax[i][0]) # Split a dataset into k folds def cross_validation_split(dataset, n_folds): dataset_split = list() dataset_copy = list(dataset) fold_size = int(len(dataset) / n_folds) for i in range(n_folds): fold = list() while len(fold) < fold_size: index = randrange(len(dataset_copy)) fold.append(dataset_copy.pop(index)) dataset_split.append(fold) return dataset_split # Calculate accuracy percentage def accuracy_metric(actual, predicted): correct = 0 for i in range(len(actual)): if actual[i] == predicted[i]: correct += 1 return correct / float(len(actual)) * 100.0 # Evaluate an algorithm using a cross validation split def evaluate_algorithm(dataset, algorithm, n_folds, *args): folds = cross_validation_split(dataset, n_folds) scores = list() for fold in folds: train_set = list(folds) train_set.remove(fold) train_set = sum(train_set, []) test_set = list() for row in fold: row_copy = list(row) test_set.append(row_copy) row_copy[-1] = None predicted = algorithm(train_set, test_set, *args) actual = [row[-1] for row in fold] accuracy = accuracy_metric(actual, predicted) scores.append(accuracy) return scores # Make a prediction with coefficients def predict(row, coefficients): yhat = coefficients[0] for i in range(len(row)-1): yhat += coefficients[i + 1] * row[i] return 1.0 / (1.0 + exp(-yhat)) # Estimate logistic regression coefficients using stochastic gradient descent def coefficients_sgd(train, l_rate, n_epoch): coef = [0.0 for i in range(len(train[0]))] for epoch in range(n_epoch): for row in train: yhat = predict(row, coef) error = row[-1] - yhat coef[0] = coef[0] + l_rate * error * yhat * (1.0 - yhat) for i in range(len(row)-1): coef[i + 1] = coef[i + 1] + l_rate * error * yhat * (1.0 - yhat) * row[i] return coef # Linear Regression Algorithm With Stochastic Gradient Descent def logistic_regression(train, test, l_rate, n_epoch): predictions = list() coef = coefficients_sgd(train, l_rate, n_epoch) for row in test: yhat = predict(row, coef) yhat = round(yhat) predictions.append(yhat) return(predictions) # Test the logistic regression algorithm on the diabetes dataset seed(1) # load and prepare data filename = 'pima-indians-diabetes.csv' dataset = load_csv(filename) for i in range(len(dataset[0])): str_column_to_float(dataset, i) # normalize minmax = dataset_minmax(dataset) normalize_dataset(dataset, minmax) # evaluate algorithm n_folds = 5 l_rate = 0.1 n_epoch = 100 scores = evaluate_algorithm(dataset, logistic_regression, n_folds, l_rate, n_epoch) print('Scores: %s' % scores) print('Mean Accuracy: %.3f%%' % (sum(scores)/float(len(scores)))) |
A k value of 5 was used for cross-validation, giving each fold 768/5 = 153.6 or just over 150 records to be evaluated upon each iteration. A learning rate of 0.1 and 100 training epochs were chosen with a little experimentation.
You can try your own configurations and see if you can beat my score.
Running this example prints the scores for each of the 5 cross-validation folds, then prints the mean classification accuracy.
We can see that the accuracy is about 77%, higher than the baseline value of 65% if we just predicted the majority class using the Zero Rule Algorithm.
|
1 2 |
Scores: [73.8562091503268, 78.43137254901961, 81.69934640522875, 75.81699346405229, 75.81699346405229] Mean Accuracy: 77.124% |
Extensions
This section lists a number of extensions to this tutorial that you may wish to consider exploring.
- Tune The Example. Tune the learning rate, number of epochs and even data preparation method to get an improved score on the dataset.
- Batch Stochastic Gradient Descent. Change the stochastic gradient descent algorithm to accumulate updates across each epoch and only update the coefficients in a batch at the end of the epoch.
- Additional Classification Problems. Apply the technique to other binary (2 class) classification problems on the UCI machine learning repository.
Did you explore any of these extensions?
Let me know about it in the comments below.
Review
In this tutorial, you discovered how to implement logistic regression using stochastic gradient descent from scratch with Python.
You learned.
- How to make predictions for a multivariate classification problem.
- How to optimize a set of coefficients using stochastic gradient descent.
- How to apply the technique to a real classification predictive modeling problem.
Do you have any questions?
Ask your question in the comments below and I will do my best to answer.







Here is a great paper describing all the different flavours of logistic regression
http://research.microsoft.com/en-us/um/people/minka/papers/logreg/
Great, thanks for the link Phil.
The above link appears to be broken. The correct link appears to now be:
https://tminka.github.io/papers/logreg/minka-logreg.pdf
Thanks Michael.
Hi Jason,
The code in 3.Diabetes Prediction throws an “ValueError: empty range for randrange()”.
I figured it only occurs when the fold_size isn’t integer, because then the “while len(fold) < fold_size" loop gets an extra iteration for each n_folds and the len(dataset_copy) eventually results to 0, throwing randrange() into an error (line 47). Changed the n_folds to 6 and it works fine. Is it something that I'm doing wrong, or the data set changed since this publication, or there is something else?
Thank you!
PS: I'm using Anaconda's Spyder 3.0.0 (Python 3.5)
Thanks Pencho, I think a cast is needed when using Python 3. I’ll make a ticket and update the example soon.
Under the cross_validation function, add a second forward slash to “fold_size = len(dataset) // n_folds”.
I have updated the cross_validation_split() function in the above example to address issues with Python 3.
Hi,
I’m curious to know why the coefficients are updated using :
coef[i + 1] = coef[i + 1] + l_rate * error * yhat * (1.0 – yhat) * row[i]
and not simply:
coef[i + 1] = coef[i + 1] + l_rate * error * row[i].
Thanks
Hi Habo,
I sourced the equation from Formula (18.8) on page 727 of Artificial Intelligence a Modern Approach.
Hi Jason,
When making the initial predictions before we optimize the coefficients, how exactly did you choose your initial coefficients? Is it just trial and error? Also, if we choose very poor coefficients (for whatever reason), can we just run the stochastic gradient algorithm to get the optimal (enough) coefficients?
Thanks
Hi Kristopher, normally we can start with zero or random coefficients.
It is good practice to re-run a stochastic process like gradient descent multiple times and keep the best outcome.
Thank you,
Unfortunately I did not see your reply until after I had asked my second question, so I apologize if the way its written seems to ignore context, I thought my initial question failed to submit.
Thanks again
Not a problem, this is a place of learning.
Hi Jason,
Why are we updating the i+1th coefficient using the ith coefficient? Why do we not just update the current coefficient we are on? In other words, why do we do:
coef[i + 1] = coef[i+1]…. etc
instead of doing
coef[i] = coef[i]…. etc
Good question Kristopher.
Because the coefficient at position 0 is the bias (intercept) coefficient which bumps the indices down one and misaligns them with the indices in the input data.
Does that make sense?
Hi Jason,
Thanks for this great post! I initially had the same thought as Habo. I wasn’t sure why it was written this way, coef[i + 1] = coef[i + 1] + l_rate * error * yhat * (1.0 – yhat) * row[i]. I actually have the AI book you referenced earlier. I took a closer look and, to me, the author is using the cost function for linear regression and substituting logistic function into h.
On the other hand, I think most logistic regression cost/loss function is written as maximum log-likelihood, which is written differently than (y – h(x))^2. Would that explain the gradient you are using in this post? It’s the difference is taking the derivative of the maximum log-likelihood versus taking the derivative of squared difference of y and estimated y? Thanks again!
Hi Habo,
I believe the terms yhat * (1.0 – yhat) are included in the estimate due to the nature of the sigmoid function and its first derivative, and trying to minimize this. The updated coefficients use these terms on each iteration for the new estimate. Think of yhat = 1 and yhat = 0. This term will disappear. So coef[i + 1] = coef[i + 1] + l_rate * error * yhat * (1.0 – yhat) * row[i] becomes coef[i + 1] = coef[i + 1]. This experimental estimate matches the actual.
I am applying the above code on my data set of ball-bearing classification problem .
At step 2 while finding the coefficients value my error it not getting reduced and its value quite high.i tried for different values of epcon and learning rate .
Consider normalizing your data and confirming that it has a Gaussian distribution.
Consider using this process to systematically work through your problem:
https://machinelearningmastery.com/start-here/#process
Thanks very much for this great post. In both examples here (both with the fake and the real data), I fit a logistic regression using the coefficients_sgd(). I also used scikit learn to fit a logistic regression. surprisingly, coefficient estimates are very different between the two approached. What would you think ha caused this?
Small differences in the implementations will cause differences in the coefficients.
There are no “best” models in applied machine learning, just lots of different models from which we must choose among:
https://machinelearningmastery.com/a-data-driven-approach-to-machine-learning/
Thanks for your answer Jason. I actually simulated data with known coefficient values. I then once fit a logistic regression with sk-learn and another time with SGD approach proposed here. The coef estimates from sk-learn are very close to the true coef values (I know the true values because I myself simulated them). However, coef estimates from SGD are very different. Could be an error in the gradient formula?
To double check the gradient formula used in the tutorial, I wrote down the loss function and took the derivatives. My derivative for coefficients (including intercept) is:
– X_i (Y_i – \hat{Y_i}) = – X_i (error)
for intercept, X_i is going to be 1.0!
Your formula for coefficients has additional elements to it. Did I make any mistake in my derivation? I modified the function and I got pretty good outputs. What would you think?
Thanks very much for your help in advance.
Errors will come from the SGD process itself and the random initial conditions.
I think regularization would be the key here. The sk-learn library does L2 regularization by default which is not done here.
I have a few basic questions :
1) Is stochastic gradient descent the only way of determining the weights/parameters, are there other ways? Please give a good resource on them.
2) When we normalize or standardize a data set and build our model on this rescaled data, what happens when theres a new data(unseen) and we try to predict using our model because imagine online streaming data we cannot determine the min/max so the new data cannot not be rescaled. So, is it like we build a model on rescaled/normalized data and predict on raw data?
No, SGD is not the only way, you can use linear algebra.
We must estimate the min/max values for scaling that take into account all new data in the future.
A good alternative is to use standardization that requires an estimate of mean and stdev rather than min/max values.
Hi Jason. I’m from Mauritius and I wanted to thank you very much for your very informative blog. I have a question on the formula you used to update your coefficients:
b1(t+1) = b1(t) + delta , where delta=learning_rate * (y(t) – yhat(t)) * yhat(t) * (1 – yhat(t)) * x1(t)
I’m trying to use the same formula for an on-line instead of a batch process but I run into the following problem:
If y(t)=0 and yhat(t)=1, delta is 0.
If y(t)=1 and yhat(t)=0, delta is 0.
These incorrect predictions get disregarded instead of updating the coefficients to try and correct them .
So my question was if the same algorithm can be used for on-line learning(i.e updating after each prediction)?
If yes am I missing something that the algorithm does or does the algorithm really disregard large errors?
Hi Jason,
When I try to run the coefficient on my own dataset, the portion (exp(-(yhat))) returns the error ‘OverflowError: math range error.’ I am using Python 3. Any ideas of where this could be coming from?
Also, one more question.
It looks like you are taking exp(-yhat), isn’t yhat more than a real number though, namely a list? How does Python handle e^(list of numbers)?
yhat is a result of a+bx (or dot product of X and coefficients), it is always a number cant be a list
Sorry to hear that, the example was developed with Python 2, perhaps it is a Python 3 issue?
Hi,
Is it possible you refer me to the derivation of this equation :
b = b + learning_rate * (y – yhat) * yhat * (1 – yhat) * x
That would be very useful.
Thanks.
I think I took it from the textbook AI a modern approah 3rd edition.
HI Jason,
I have implemented the above method on a demo data set of my own.
However, when i do this, the SGD model produced (after 10 epochs of the data) does not have coefficients that are anywhere near the coefficients produced from building a traditional logistic model using the IRLS method on this demo data-set. I would expect the coefficients of the SGD model to at least be converging towards the ‘true” coefficient values from the traditional IRLS model.
Do you have any ideas as to why this may be the case? Could it be because the dataset i am using is ordered ? At the moment all of my ‘good acocunts (Y=1) come first then all my ‘bad’ accounts (Y=0) come after. Should i randomize the dataset before applying SGD?
Also – the final coefficients for the SGD model after 10 eopchs seem to be utterly dependant on the ‘learning rate’ and, from what i can make out, there is no ‘correct’ value to use here. Can you help me?
Hi Alex, the SGD is probably getting stuck in local optima, for small problems using a linalg solution is more efficient and accurate. SGD will need tuning.
Hi Jason,
Thanks for getting back to me. I have a couple more questions if you don’t mind:
1) What is a linalg solution?
2) I have also built a traditional logistic model (IRLS) on a small sample of the dataset and then trained this model on the remaining data. I thought that this would mean the ‘start point’ for SGD is already close to the global optima and would therefore work better but i am still having no luck here – this SGD model is still nowehere near the traditional IRLS model built on the entire dataset – do you have an idea why this may be happening?
Thanks again for your assistance
By linalg I mean estimating the solution directly using matric operations (analytically), rather than searching for a solution (optimization).
Some ideas:
– Maybe the SGD needs tuning on your problem.
– Maybe an analytical solution is the best for your problem.
– Maybe there is a bug in the code.
Hello Jason, your article on SGD in logistic regression was very helpful. I am trying to implement the SGD algorithm, but I have a question. How should I handle variables that are categorical in nature ? Should I simply treat them as numeric variables and have the SGD algorithm estimates coefficients for them ?
Thanks in advance !
Generally, I would recommend converting them to numeric or binary vectors (one hot encoded).
Thanks for the quick reply ! I actually ended finding the answer in this very blog not long after I asked.
I ran the example on my machine with the diabetes dataset and, as expected, it gave a precision of ~77%. When I ran it with the “Logistic” classifier under Weka, it gave a similar result but with coefficients that were vastly different from the ones I got after running the code posted above. The coefficients Weka gave me stay the same regardless of what boolean value I give to the useConjugateGradientDescent option.
I then re-ran it using the SGD qualifier (with and without normalization) this time, only to get yet another different set of coefficients, albeit with the same precision.
Is it normal for the coefficients to differ depending on which algorithm we use ?
Yes.
Hi, Jason
The Line 72 “row_copy[-1] = None” is necessary? What’s for?
Many thanks!
To clear the results so the algorithm cannot “cheat” intentionally or by a bug.
Great!
Better to move it forward 1 line?
No need, we still have a reference to the list so that we can change values within it.
Hi Jason, nice post. Is there a reason why there is no convergence criteria set when learning parameter vector theta so that there is no need to iterate over all epochs?
Thanks,
Dan
To keep the example simple. Please modify it to add any criteria you wish.
Hi Jason,
Thanks for posting really nice post. I have a questions, why did you choose the MSE cost function rather than cross entropy because the prediction function is non-linear due to sigmoid transform.
Thanks,
Sanchit
Perhaps try using log loss and see how you go.
Thanks. One more question, does this approach limited to number of features in the dataset?
for example, I have more than 20 features in my dataset.
the same approach should work right?
I’m sure it will. More features are more complexity.
20 is not a lot of features. Try it and see.
Thanks Jason for posting this wonderful piece of knowledge.
I don’t know why and where, but running this code on Python 3 gives me:
Scores: [9.15032679738562, 4.57516339869281, 11.76470588235294, 8.49673202614379, 7.18954248366013]
Mean Accuracy: 8.235%
Trying to find out…
The code was written for Py2.7.
Forget about Jason,
I figured out what was happening.
Class columns was in the first position instead of last.
Thanks
Happy to hear that.
Hi Jason,
Assuming I have a seperate array for labels, where my data looks like [12, 136, 34] and the labels [1]. Can I just use the b0 first coefficient as well? or do I have to ignore it? Because i can see from your data that the last column is the label if I am not mistaken
Sorry, I don’t follow your question. Can you elaborate?
Hey Jason,
Thanks for your excellent articles! I have a question about implementing backpropagation and optimization for the case of a mini-batch.
We will essentially have a loss per example, L_i. Most toolkits seem to return an average loss, so something like (1/n)*sum(L_i’s) where n is the number of samples in the minibatch. However, we still need to compute the gradient of coef for each example right (lets call it G_i)?
So do we then average all the gradients (across samples) and do an update? i.e. coef = coef – lr * average(G_i’s)
What is the difference between the above method, and the method where each G_i is computed with its own L_i (instead of average(L_i’s)), and the average gradient ( average(G_i’s) ) is used for the update?
Yes, we average the loss over many samples. If we don’t we get a very noisy estimate of the gradient, and in turn noisy updates to weights.
If we average the loss, we still get one set of gradients G_i for each input in the minibatch right? (Since the gradient might itself depend on the input).
So when performing the parameter updates, do we average the gradients G_i’s again? If yes, why do we need to perform averaging at two steps (loss AND gradients), and not only once for the gradients?
Hi, I noticed in your code (when doing stochastic gradient descent) for the linear regression, you had this
coef[i + 1] = coef[i + 1] – l_rate * error * row[i]
But for the logistic regression, you had this
coef[i + 1] = coef[i + 1] + l_rate * error * yhat * (1.0 – yhat) * row[i]
Is the yhat * (1.0 – yhat) part of the function equivalent to ln(y / (1-y)) ?
Good question, I’m not sure off the cuff, I don’t the derivation at hand.
can any one assist me to do this stochastic gradient descent using R will be greatful
I recommend using a library rather than coding it yourself.
How can we get the coefficients of the independent variables using python code?
I show you how to compute them and print them in the above tutorial.
Which part are you having trouble with?
Hi, Jason!
Another excellent post from you. Thank you.
I tried to be smart (or lazy) and use the Scikit-learn API for SGD Logistic Regression.
Comparing apples to apples, the API gets 63% accuracy while with your code, I get a 77% accuracy.
Strange and interesting ….
Well done!
Yes, small differences in implementation can have a large impact in performance.
Thanks Jason!
Hi jason, thanks for your interessting tutorials. I heve a question . Which is better to use for improving the accuracy and auc : SGD or Xgboost ?
Try both and see what works best for your specific dataset.
Hi Jason, I loved your implementation.
Interestingly our class often use the gradient ascent to find the coefficient W which maximize the log of conditional likelihood of the P(Y|X, W).
And I believe in this implementation we instead using the gradient descent to minimize the a cost function. I would assume the cost function is Y_true – P(Y|X, W), would you please confirm?
The log of conditional likelihood can be found on Page 11 of the following notes:
http://www.cs.cmu.edu/~tom/mlbook/NBayesLogReg.pdf
Hi Jason!
Do we have to scale and normalize the data? as far as i am aware of it is not a must in logistic regression models.
If we have to, why?
Thankyou very much!
It depends on the data, if the units differ across input variables then yes, it is advised to scale the inputs to the same range.
Try with and without scaling and compare performance.
Thanks for the answer, Jason!
Actually, I did, and for me, it was the same score.
But I will note that from now on.
thank you jason, Highly informative article.
Thanks, I’m happy it helped.
Hi sir,
i have a doubt
The coefficients obtained by LinearRegression() from sklearn.linear_model is also better coefficients right – these coefficients are obtained by minimising the OLS
Then why we are applying stochastic gradient descent again to obtain the same coefficients
the coefficients values from ML models are not accurate or not?
Or else stochastic gradient descent is used to get much better coefficients values?
There are many ways to solve the linear regression.
The linalg approach is good when all data fits into memory.
The sgd approach is good when you have a lot of data.
Both can be accurate or inaccurate, sgd can have more noise in the result but is more robust. linalg has strict assumptions about the data.
Hi Jason, here in SGD you have used square loss which is linear regression. Can you post log loss method for logistic loss.
Thanks for the suggestion.
Hi Jason,
Again great blog and great article.
Small issue, in the link you provide, the file name is “pima-indians-diabetes.data.csv” whereas in the code it is “pima-indians-diabetes.csv” so the interpreter won’t find it.
For the readers who encounter this error, just alter the names so that they match.
Thanks
Thanks for the tip.
Note, the tutorial says:
Hi Jason,
As a follow up, a couple of questions:
1- Why is this called “stochastic” gradient descent? I have the impression when I look at the code of coefficients_sgd() that it is a deterministic algorithm and running it several with the same parameters yielded the same results.Could you clarify what in this context ‘stochastic’ means?
2- You have mentioned to Manuela that the implementing it with xgboost was feasible. Would that still be a form of Logistic Regression or would that be considered a decision tree algorithm?
Thanks again for your site and all the quality material you share !
Because the update of weights after each samples is noisy – e.g. a stochastic estimate of the gradient.
XGBoost does not use SGD, but there it can use stochastic sampling of features and rows, called stochastic gradient boosting.
Hi Jason,
could you please tell me how did you choose the intial coefficients here
y = 1.0 / (1.0 + e^(-(-0.406605464 + 0.852573316 * X1 + -1.104746259 * X2))) 1 y = 1.0 / (1.0 + e^(-(-0.406605464 + 0.852573316 * X1 + -1.104746259 * X2)))
Thanks,
Mahmoud
In that case I think they were arbitrary for the test of that bit of code.
The whole idea of training a logistic regression model is to find those coefficients.
Hi Jason:
First of all, thank you very much for sharing; every article you write is great.
I have a small question to ask you: I noticed in your article about b0, b1, b2 … The iterative update formula is applied [the textbook AI a modern approah 3rd edition], I found the book and read the relevant part, and then I found that the loss function used in this part is loss (x) = y-yhat; then I searched for other maximum likelihoods used in the implementation of logistic regression stochastic gradient descent The function is (pi ^ y (1-pi) 1-y). So are these two ideas different? The idea used by the maximum likelihood function is the idea of maximum likelihood, and the idea used in your article is to minimize the error directly?
Looking forward to your reply. Thank you.
You’re welcome.
Yes, there are many approaches. Here we are learning about the structure of the model and how to use an optimization algorithm to solve it, not the optimal approach to solving logistic regression.
Also, more on MLE for LR here:
https://machinelearningmastery.com/logistic-regression-with-maximum-likelihood-estimation/
Hi Jason:
First of all, thank you very much for sharing your article which is so helpful for all Machine Learning Engineers.I am really impressed with your articles and no words to say.
All of your articles are awesome and really helping to the Datascience world.I really appreciate your efforts and contributions to all Datascience world.
It will be more helpful if you post articles on some real world projects from scrtach to end.Or let me know any links if you already post any.Or suggest me some links where I can find.
Looking forward to your reply. Thank you once again with Happy new year wishes.
You’re welcome.
Yes, there are hundreds of projects on the blog, you can use the search to find them.
Hi Jason,
I performed the steps in XL sheet,
But, i got incorrect results.
I cross checked multiple times .
where exactly i went wrong.
I followed the example given in 14.3 from the book “Master Machine Learning Algorithms”
https://drive.google.com/file/d/1jQgn4yy9DYrMWmyKyxY3VECQ1hDLsfE2/view?usp=sharing
Find the Xl sheet .
Thanking you
Please refer to the spreadsheets provided with the book and compare results.
If you have lost the spreadsheets provided with the book, email me and I can resend you purchase receipt with an updated download link:
https://machinelearningmastery.com/contact/
If you pirated the book, it would not be ethical for me to help you.
Hi Jason,
Any pointers on trying to run the gradient computation through multiprocessing
Yes, use an existing implementation that support multithreading.
Also logistic regression is not really a parallel-able algorithm, unless you change it to use SGD, then you can do coefficient updates in batches.
Dear Colleague!
b = b + learning_rate * (y – yhat) * x
because difference loss-function (cross-entropy function)
L = – sum(yhat * ln(y) + (1 – yhat) * (1 – ln(y)))
Hi Jason,
I just wanted to add to my earlier comment. I believe we should note that the gradient used in the post is the derivative of the L2 loss function, one of many loss/cost functions used to calculate the error.
On the other hand, we would get something like this, b = b + learning_rate * (y – yhat) * x, if the gradient (derivative) of the log-likelihood is taken. Thus, this gradient would be used to maximize the log-likelihood and determine the best fitting coefficients through gradient descent. Let me know what you think. Thanks!
My apologies … I should have written best fitting coefficients through “gradient ascent” because we are maximizing the log-likelihood. We are taking small steps in the direction of the gradient and reaching the local maxima.
ML guru Andrew Ng uses the derivative of log loss function when computing the stochastic gradient descent for logistic regression. I am not sure why this is not implemented in this chapter and what is the significance of this chapter’s gradient method as compared to log loss derivative approach.
I have the same question: to me, the only difference btwn this and the linear regression example from another of Jason’s examples should be the function h_theta, called “predict” here. Thus, I believe the loss function should be; coef[i + 1] = coef[i + 1] + l_rate * error * row[i] , but I’d love to know if I’m misinterpreting
You seem to be using square loss
J = (y-y^)**2
dJ/db1 = dJ/dy * dy/db1
= – 2 * (y-y^) * (y^) (1-y^)* x
In which case you missed out “2” , Jason am i correct ??
Correct but do you see that’s no difference from the gradient descent perspective?
Hi,
I don’t understand in the equation b = b + learning_rate * (y – yhat) * yhat * (1 – yhat) * x, wich x is this? x1 or x2?
Hi Andrew…Should be X1 in this case.
Hi Jason, I don’t understand meaning of normalize_dataset() function
Hi cylon…The following may be of interest to you:
https://machinelearningmastery.com/batch-normalization-for-training-of-deep-neural-networks/
why calculate like this row[i] = (row[i] – minmax[i][0]) / (minmax[i][1] – minmax[i][0]),Is it not possible to use the dataset itself?
Hi cylon…Yes, there are other options. How would you approach it differently?
Hi Jason,
I have a basic questions.
On the final output for the real dataset, we only get Mean Accuracy: 77.124%, but do not print the optimal parameters.
May I ask how to print the set of coefficients associated with Mean Accuracy: 77.124%?