In this post you will discover 7 recipes for non-linear classification with decision trees in R.
All recipes in this post use the iris flowers dataset provided with R in the datasets package. The dataset describes the measurements if iris flowers and requires classification of each observation to one of three flower species.
Kick-start your project with my new book Machine Learning Mastery With R, including step-by-step tutorials and the R source code files for all examples.
Let’s get started.

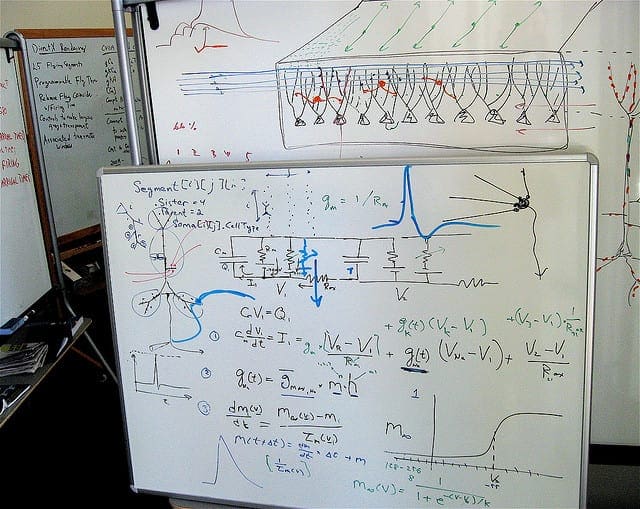
Classification with Decision Trees
Photo by stwn, some rights reserved
Classification and Regression Trees
Classification and Regression Trees (CART) split attributes based on values that minimize a loss function, such as sum of squared errors.
The following recipe demonstrates the recursive partitioning decision tree method on the iris dataset.
|
1 2 3 4 5 6 7 8 9 10 11 12 |
# load the package library(rpart) # load data data(iris) # fit model fit <- rpart(Species~., data=iris) # summarize the fit summary(fit) # make predictions predictions <- predict(fit, iris[,1:4], type="class") # summarize accuracy table(predictions, iris$Species) |
Learn more about the rpart function and the rpart package.
C4.5
The C4.5 algorithm is an extension of the ID3 algorithm and constructs a decision tree to maximize information gain (difference in entropy).
The following recipe demonstrates the C4.5 (called J48 in Weka) decision tree method on the iris dataset.
|
1 2 3 4 5 6 7 8 9 10 11 12 |
# load the package library(RWeka) # load data data(iris) # fit model fit <- J48(Species~., data=iris) # summarize the fit summary(fit) # make predictions predictions <- predict(fit, iris[,1:4]) # summarize accuracy table(predictions, iris$Species) |
Learn more about the J48 function and the RWeka package.
Need more Help with R for Machine Learning?
Take my free 14-day email course and discover how to use R on your project (with sample code).
Click to sign-up and also get a free PDF Ebook version of the course.
PART
PART is a rule system that creates pruned C4.5 decision trees for the data set and extracts rules and those instances that are covered by the rules are removed from the training data. The process is repeated until all instances are covered by extracted rules.
The following recipe demonstrates the PART rule system method on the iris dataset.
|
1 2 3 4 5 6 7 8 9 10 11 12 |
# load the package library(RWeka) # load data data(iris) # fit model fit <- PART(Species~., data=iris) # summarize the fit summary(fit) # make predictions predictions <- predict(fit, iris[,1:4]) # summarize accuracy table(predictions, iris$Species) |
Learn more about the PART function and the RWeka package.
Bagging CART
Bootstrapped Aggregation (Bagging) is an ensemble method that creates multiple models of the same type from different sub-samples of the same dataset. The predictions from each separate model are combined together to provide a superior result. This approach has shown participially effective for high-variance methods such as decision trees.
The following recipe demonstrates bagging applied to the recursive partitioning decision tree for the iris dataset.
|
1 2 3 4 5 6 7 8 9 10 11 12 |
# load the package library(ipred) # load data data(iris) # fit model fit <- bagging(Species~., data=iris) # summarize the fit summary(fit) # make predictions predictions <- predict(fit, iris[,1:4], type="class") # summarize accuracy table(predictions, iris$Species) |
Learn more about the bagging function and the ipred package.
Random Forest
Random Forest is variation on Bagging of decision trees by reducing the attributes available to making a tree at each decision point to a random sub-sample. This further increases the variance of the trees and more trees are required.
The following recipe demonstrate the random forest method applied to the iris dataset.
|
1 2 3 4 5 6 7 8 9 10 11 12 |
# load the package library(randomForest) # load data data(iris) # fit model fit <- randomForest(Species~., data=iris) # summarize the fit summary(fit) # make predictions predictions <- predict(fit, iris[,1:4]) # summarize accuracy table(predictions, iris$Species) |
Learn more about the randomForest function and the randomForest package.
Gradient Boosted Machine
Boosting is an ensemble method developed for classification for reducing bias where models are added to learn the misclassification errors in existing models. It has been generalized and adapted in the form of Gradient Boosted Machines (GBM) for use with CART decision trees for classification and regression.
The following recipe demonstrate the Gradient Boosted Machines (GBM) method in the iris dataset.
|
1 2 3 4 5 6 7 8 9 10 11 12 |
# load the package library(gbm) # load data data(iris) # fit model fit <- gbm(Species~., data=iris, distribution="multinomial") # summarize the fit print(fit) # make predictions predictions <- predict(fit, iris) # summarize accuracy table(predictions, iris$Species) |
Learn more about the gbm function and the gbm package.
Boosted C5.0
The C5.0 method is a further extension of C4.5 and pinnacle of that line of methods. It was proprietary for a long time, although the code was released recently and is available in the C50 package.
The following recipe demonstrates the C5.0 with boosting method applied to the iris dataset.
|
1 2 3 4 5 6 7 8 9 10 11 12 |
# load the package library(C50) # load data data(iris) # fit model fit <- C5.0(Species~., data=iris, trials=10) # summarize the fit print(fit) # make predictions predictions <- predict(fit, iris) # summarize accuracy table(predictions, iris$Species) |
Learn more about the C5.0 function in the C50 package.
Summary
In this post you discovered 7 recipes for non-linear classification using decision trees in R using the iris flowers dataset.
Each recipe is generic and ready for you to copy and paste and modify for your own problem.





Hi Jason,
I have a question that is how to classifier land use type with remotely sensed data using decision tree in R. Could you give me some advice? Thank you very much.
Jimmy
Hi,
I have very less knowledge about R. Recently I am trying to do rule based classification and i found that R has C50 package that can do the task.
I want to ask if it possible to optimize the number of rule in this package by adding some optimization coding or package?
Thank you in advance.
Best regards,
Grace
Thank you for your post…I tried to reproduce each scenario. The best result is with C5.0. I have just a comment. I could not reproduce the result under GBM. I get an error at the line: predictions <- predict(fit, iris)
Error in paste("Using", n.trees, "trees…\n") :
argument "n.trees" is missing, with no default
Regards,
David
I got the same error. I found that putting in the argument n.trees=iris$Species fixes that error. Now I am getting an error that says Error in if (missing(n.trees) || any(n.trees > object$n.trees)) { :
missing value where TRUE/FALSE needed.
I’m trying to find what all of the arguments for predict are for gbm, but that is proving difficult.
Hi,
I am working with high unbalanced data.
could you introduce me some methods for handling imbalanced data in R?
Bests
Thanks for the suggestion.
Love all your pages! Parts of the codes dont work well 🙁 especially with predict(). With GBM type = “class” is not allowed and therefore getting the predicted class is not provided. This has the consequence that no table or confusionmatrix can be made… Is there a package that i miss or something?
Keep up the good work!!
Thanks.
Are you able to confirm that your packages are installed and updated to the latest versions?