The data features that you use to train your machine learning models have a huge influence on the performance you can achieve.
Irrelevant or partially relevant features can negatively impact model performance.
In this post you will discover automatic feature selection techniques that you can use to prepare your machine learning data in python with scikit-learn.
Kick-start your project with my new book Machine Learning Mastery With Python, including step-by-step tutorials and the Python source code files for all examples.
Let’s get started.
- Update Dec/2016: Fixed a typo in the RFE section regarding the chosen variables.
- Update Mar/2018: Added alternate link to download the dataset.
- Update Sep/2019: Fixed code to be compatible with Python 3.
- Update Dec/2019: Updated univariate selection to use ANOVA.

Feature Selection For Machine Learning in Python
Photo by Baptiste Lafontaine, some rights reserved.
Feature Selection
Feature selection is a process where you automatically select those features in your data that contribute most to the prediction variable or output in which you are interested.
Having irrelevant features in your data can decrease the accuracy of many models, especially linear algorithms like linear and logistic regression.
Three benefits of performing feature selection before modeling your data are:
- Reduces Overfitting: Less redundant data means less opportunity to make decisions based on noise.
- Improves Accuracy: Less misleading data means modeling accuracy improves.
- Reduces Training Time: Less data means that algorithms train faster.
You can learn more about feature selection with scikit-learn in the article Feature selection.
Need help with Machine Learning in Python?
Take my free 2-week email course and discover data prep, algorithms and more (with code).
Click to sign-up now and also get a free PDF Ebook version of the course.
Feature Selection for Machine Learning
This section lists 4 feature selection recipes for machine learning in Python
This post contains recipes for feature selection methods.
Each recipe was designed to be complete and standalone so that you can copy-and-paste it directly into you project and use it immediately.
Recipes uses the Pima Indians onset of diabetes dataset to demonstrate the feature selection method . This is a binary classification problem where all of the attributes are numeric.
1. Univariate Selection
Statistical tests can be used to select those features that have the strongest relationship with the output variable.
The scikit-learn library provides the SelectKBest class that can be used with a suite of different statistical tests to select a specific number of features.
Many different statistical test scan be used with this selection method. For example the ANOVA F-value method is appropriate for numerical inputs and categorical data, as we see in the Pima dataset. This can be used via the f_classif() function. We will select the 4 best features using this method in the example below.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 |
# Feature Selection with Univariate Statistical Tests from pandas import read_csv from numpy import set_printoptions from sklearn.feature_selection import SelectKBest from sklearn.feature_selection import f_classif # load data filename = 'pima-indians-diabetes.data.csv' names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class'] dataframe = read_csv(filename, names=names) array = dataframe.values X = array[:,0:8] Y = array[:,8] # feature extraction test = SelectKBest(score_func=f_classif, k=4) fit = test.fit(X, Y) # summarize scores set_printoptions(precision=3) print(fit.scores_) features = fit.transform(X) # summarize selected features print(features[0:5,:]) |
For help on which statistical measure to use for your data, see the tutorial:
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
You can see the scores for each attribute and the 4 attributes chosen (those with the highest scores). Specifically features with indexes 0 (preq), 1 (plas), 5 (mass), and 7 (age).
|
1 2 3 4 5 6 7 |
[ 39.67 213.162 3.257 4.304 13.281 71.772 23.871 46.141] [[ 6. 148. 33.6 50. ] [ 1. 85. 26.6 31. ] [ 8. 183. 23.3 32. ] [ 1. 89. 28.1 21. ] [ 0. 137. 43.1 33. ]] |
2. Recursive Feature Elimination
The Recursive Feature Elimination (or RFE) works by recursively removing attributes and building a model on those attributes that remain.
It uses the model accuracy to identify which attributes (and combination of attributes) contribute the most to predicting the target attribute.
You can learn more about the RFE class in the scikit-learn documentation.
The example below uses RFE with the logistic regression algorithm to select the top 3 features. The choice of algorithm does not matter too much as long as it is skillful and consistent.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 |
# Feature Extraction with RFE from pandas import read_csv from sklearn.feature_selection import RFE from sklearn.linear_model import LogisticRegression # load data url = "https://raw.githubusercontent.com/jbrownlee/Datasets/master/pima-indians-diabetes.csv" names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class'] dataframe = read_csv(url, names=names) array = dataframe.values X = array[:,0:8] Y = array[:,8] # feature extraction model = LogisticRegression(solver='lbfgs') rfe = RFE(model, 3) fit = rfe.fit(X, Y) print("Num Features: %d" % fit.n_features_) print("Selected Features: %s" % fit.support_) print("Feature Ranking: %s" % fit.ranking_) |
You can see that RFE chose the the top 3 features as preg, mass and pedi.
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
These are marked True in the support_ array and marked with a choice “1” in the ranking_ array.
|
1 2 3 |
Num Features: 3 Selected Features: [ True False False False False True True False] Feature Ranking: [1 2 3 5 6 1 1 4] |
3. Principal Component Analysis
Principal Component Analysis (or PCA) uses linear algebra to transform the dataset into a compressed form.
Generally this is called a data reduction technique. A property of PCA is that you can choose the number of dimensions or principal component in the transformed result.
In the example below, we use PCA and select 3 principal components.
Learn more about the PCA class in scikit-learn by reviewing the PCA API. Dive deeper into the math behind PCA on the Principal Component Analysis Wikipedia article.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 |
# Feature Extraction with PCA import numpy from pandas import read_csv from sklearn.decomposition import PCA # load data url = "https://raw.githubusercontent.com/jbrownlee/Datasets/master/pima-indians-diabetes.csv" names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class'] dataframe = read_csv(url, names=names) array = dataframe.values X = array[:,0:8] Y = array[:,8] # feature extraction pca = PCA(n_components=3) fit = pca.fit(X) # summarize components print("Explained Variance: %s" % fit.explained_variance_ratio_) print(fit.components_) |
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
You can see that the transformed dataset (3 principal components) bare little resemblance to the source data.
|
1 2 3 4 5 6 7 |
Explained Variance: [ 0.88854663 0.06159078 0.02579012] [[ -2.02176587e-03 9.78115765e-02 1.60930503e-02 6.07566861e-02 9.93110844e-01 1.40108085e-02 5.37167919e-04 -3.56474430e-03] [ 2.26488861e-02 9.72210040e-01 1.41909330e-01 -5.78614699e-02 -9.46266913e-02 4.69729766e-02 8.16804621e-04 1.40168181e-01] [ -2.24649003e-02 1.43428710e-01 -9.22467192e-01 -3.07013055e-01 2.09773019e-02 -1.32444542e-01 -6.39983017e-04 -1.25454310e-01]] |
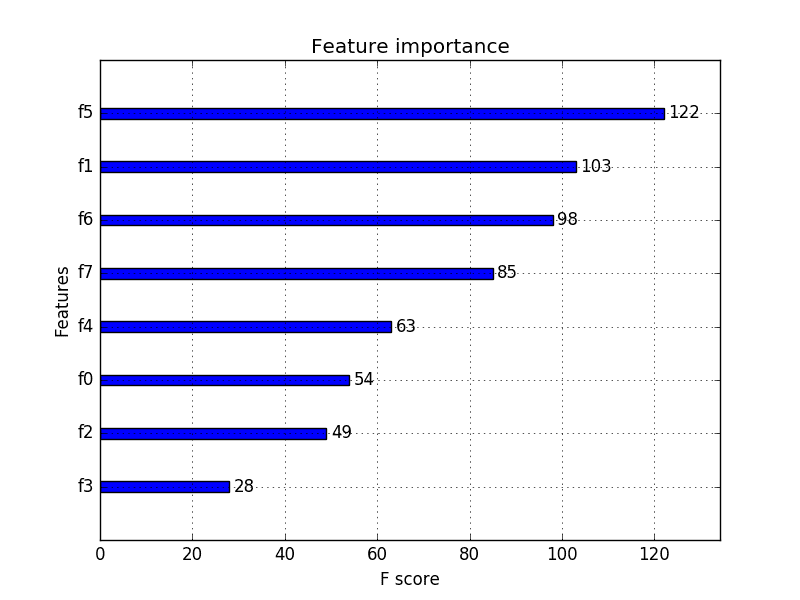
4. Feature Importance
Bagged decision trees like Random Forest and Extra Trees can be used to estimate the importance of features.
In the example below we construct a ExtraTreesClassifier classifier for the Pima Indians onset of diabetes dataset. You can learn more about the ExtraTreesClassifier class in the scikit-learn API.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 |
# Feature Importance with Extra Trees Classifier from pandas import read_csv from sklearn.ensemble import ExtraTreesClassifier # load data url = "https://raw.githubusercontent.com/jbrownlee/Datasets/master/pima-indians-diabetes.csv" names = ['preg', 'plas', 'pres', 'skin', 'test', 'mass', 'pedi', 'age', 'class'] dataframe = read_csv(url, names=names) array = dataframe.values X = array[:,0:8] Y = array[:,8] # feature extraction model = ExtraTreesClassifier(n_estimators=10) model.fit(X, Y) print(model.feature_importances_) |
Note: Your results may vary given the stochastic nature of the algorithm or evaluation procedure, or differences in numerical precision. Consider running the example a few times and compare the average outcome.
You can see that we are given an importance score for each attribute where the larger score the more important the attribute. The scores suggest at the importance of plas, age and mass.
|
1 |
[ 0.11070069 0.2213717 0.08824115 0.08068703 0.07281761 0.14548537 0.12654214 0.15415431] |
Summary
In this post you discovered feature selection for preparing machine learning data in Python with scikit-learn.
You learned about 4 different automatic feature selection techniques:
- Univariate Selection.
- Recursive Feature Elimination.
- Principle Component Analysis.
- Feature Importance.
If you are looking for more information on feature selection, see these related posts:
- Feature Selection with the Caret R Package
- Feature Selection to Improve Accuracy and Decrease Training Time
- An Introduction to Feature Selection
- Feature Selection in Python with Scikit-Learn
Do you have any questions about feature selection or this post? Ask your questions in the comment and I will do my best to answer them.




Hi Jason! Thanks for this – really useful post! I’m sure I’m just missing something simple, but looking at your Univariate Analysis, the features you have listed as being the most correlated seem to have the highest values in the printed score summary. Is that just a quirk of the way this function outputs results? Thanks again for a great access-point into feature selection.
Hi Juliet, it might just be coincidence. If you uncover something different, please let me know.
It is sound good .
Does deep learning need feature selection?
Thank you
It may need. The best way to tell is to see if feature selection can improve the result.
Yes, Right, feature selection will improve the overall result.
Absolutely Alok! Keep up the great work!
For the Recursive Feature Elimination, are the features of high importance (preg,mass,pedi)?
The ranking array has value 1 for them them.
Hi Ansh, I believe the features with the 1 are preg, pedi and age as mentioned in the post. These are the first ranked features.
Thanks for the reply Jason. I seem to have made a mistake, my bad. Great post 🙂
No problem Ansh.
Hi all,
I agree with Ansh. There are 8 features and the indexes with True and 1 match with preg, mass and pedi.
[ ‘preg’, ‘plas’, ‘pres’, ‘skin’, ‘test’, ‘mass’, ‘pedi’, ‘age’ ]
[ True, False, False, False, False, True, True, False]
[ 1, 2, 3, 5, 6, 1, 1, 4 ]
Jason, could you explain better how you see that preg, pedi and age are the first ranked features?
Thank you for the post, it was very useful and direct to the point. Congratulations.
Hi Anderson, they have a “true” in their column index and are all ranked “1” at their respective column index.
Does that help?
Hi Jason,
That is exactly what I mean. I believe that the best features would be preg, pedi and age in the scenario below
Features:
[ ‘preg’, ‘plas’, ‘pres’, ‘skin’, ‘test’, ‘mass’, ‘pedi’, ‘age’ ]
RFE result:
[ True, False, False, False, False, False, True, True ]
[ 1, 2, 3, 5, 6, 4, 1, 1 ]
However, the result was
Features:
[ ‘preg’, ‘plas’, ‘pres’, ‘skin’, ‘test’, ‘mass’, ‘pedi’, ‘age’ ]
RFE result:
[ True, False, False, False, False, True, True, False]
[ 1, 2, 3, 5, 6, 1, 1, 4 ]
Did you consider the target column ‘class’ by mistake?
Thank you for the quick reply,
Anderson Neves
Hi Anderson,
I see, you’re saying you have a different result when you run the code?
The code is correct and does not include the class as an input.
Re-running now I see the same result:
Perhaps I don’t understand the problem you’ve noticed?
Hi Jason,
Your code is correct and my result is the same as yours. My point is that the best features found with RFE are preg, mass and pedi. So, I suggest you fix the text “You can see that RFE chose the the top 3 features as preg, pedi and age.”. If you add the code below at the end of your code you will see what I mean.
# find best features
best_features = []
i = 0
for is_best_feature in fit.support_:
if is_best_feature:
best_features.append(names[i])
i += 1
print ‘\nSelected features:’
print best_features
Sorry if I am bothering somehow,
Thanks again,
Anderson Neves
Got it Anderson.
Thanks for being patient with me and helping to make this post more useful. I really appreciate it!
I’ve fixed up the example above.
Hi, thank you for this post, can I use theses selected features algorithm for (knn, svm, dicision tree, logic regression)? For example, RFE are used only with logic regression or I can use with any classification algorithm?
You can use any algorithm, see this:
https://machinelearningmastery.com/rfe-feature-selection-in-python/
from the rfe, how do I form a new dataframe for the features which has true value?
Great question Narasimman.
From memory, you can use numpy.concatinate() to collect the columns you want.
http://docs.scipy.org/doc/numpy/reference/generated/numpy.concatenate.html
Thanks for useful tutorial.
Narasimman – ‘from the rfe, how do I form a new dataframe for the features which has true value?’
You can just apply rfe directly to the dataframe then select based on columns:
…
df = read_csv(url, names=names)
X = df.iloc[:, 0:8]
Y = df.iloc[:, 8]
# feature extraction
model = LogisticRegression()
rfe = RFE(model, 3)
fit = rfe.fit(X, Y)
print(“Num Features: {}”.format(fit.n_features_))
print(“Selected Features: {}”.format(fit.support_))
print(“Feature Ranking: {}”.format(fit.ranking_))
X = X[X.columns[fit.support_]]
Hi Jason,
Really appreciate your post! Really great! I have a quick question for the PCA method. How to get the column header for the selected 3 principal components? It is just simple column no. there, but hard to know which attributes finally are.
Thanks,
Thanks MLBeginner, I’m glad you found it useful.
There is no column header, they are “new” features that summarize the data. I hope that helps.
hi, Jason! please I want to ask you if i can use PSO for feature selection in sentiment analysis by python
Sure, try it and see how the results compare (as in the models trained on selected features) to other feature selection methods.
Hey Jason, can the univariate test of Chi2 feature selection be applied to both continuous and categorical data.
Hi Vignesh, I believe just continuous data. But I may be wrong – try and see.
Hey Jason, Thanks for the reply. In the univariate selection to perform the chi-square test you are fetching the array from df.values. In that case, each element of the array will be each row in the data frame.
To perform feature selection, we should have ideally fetched the values from each column of the dataframe to check the independence of each feature with the class variable. Is it a inbuilt functionality of the sklearn.preprocessing beacuse of which you fetch the values as each row.
Please suggest me on this.
I’m not sure I follow Vignesh. Generally, yes, we are using built-in functions to perform the tests.
Hi Jason,
I am trying to do image classification using cpu machine, I have very large training matrix of 3800*200000 means 200000 features. Pls suggest how do I reduce my dimension.?
Consider working with a sample of the dataset.
Consider using the feature selection methods in this post.
Consider projection methods like PCA, sammons mapping, etc.
I hope that helps as a start.
Hi Jason:
import numpy as np
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import chi2
most_relevant = SelectKBest(chi2, k>=4).fit(X_train, y_train)
most_relevant_df = pd.DataFrame(zip(X_train.columns, most_relevant.scores_),
columns= [‘Variables’, ‘score’]).sort_values( ‘score’, ascending=False).head(20)
most_relevant_variables = most_relevant_df.Variables.tolist()
most_relevant_df
—————————————————————————
NameError Traceback (most recent call last)
in
2 from sklearn.feature_selection import SelectKBest
3 from sklearn.feature_selection import chi2
—-> 4 most_relevant = SelectKBest(chi2, k>=4).fit(X_train, y_train)
5 most_relevant_df = pd.DataFrame(zip(X_train.columns, most_relevant.scores_),
6 columns= [‘Variables’, ‘score’]).sort_values( ‘score’, ascending=False).head(20)
NameError: name ‘k’ is not defined
i am having this issue, K not defined, how or i need in the past i have use this code and no need, do you know what can be ?
This will help you copy the code correctly:
https://machinelearningmastery.com/faq/single-faq/how-do-i-copy-code-from-a-tutorial
Jason,
when you use “SelectKBest” , can you please explain how you get the below scores?
[ 111.52 1411.887 17.605 53.108 2175.565 127.669 5.393
181.304]
-Mani
I use a chi squared test, you can learn more about it here:
http://scikit-learn.org/stable/modules/generated/sklearn.feature_selection.chi2.html#sklearn.feature_selection.chi2
Jason,
I understand you used chi square. But if want to get these scores manually , how can i do it? Please explain.
-Mani
Good question, I don’t have an example at the moment sorry.
jason,
Please explain how the below scores are achieved using chi2.
[ 111.52 1411.887 17.605 53.108 2175.565 127.669 5.393
181.304]
-Mani
Jason, how can we get feature names from their rankings?
Hi Natheer,
Map the feature rank to the index of the column name from the header row on the DataFrame or whathaveyou.
Hi Jason,
Thank you for this nice blog
I have a regression problem and I need to convert a bunch of categorical variables into dummy data, which will generate over 200 new columns. Should I do the feature selection before this step or after this step?
Thanks
Try and see.
That is a lot of new binary variables. Your resulting dataset will be sparse (lots of zeros). Feature selection prior might be a good idea, also try after.
Hi Jason,
I am bit stuck in selecting the appropriate feature selection algorithm for my data.
I have about 900 attributes (columns) in my data and about 60 records. The values are nothing but count of attributes.
Basically, I am taking count of API calls of a portable file.
My data is like this:
File, dangerous, API 1,API 2,API 3,API 4,API 5,API 6…..API 900
ABC, yes, 1,0,2,1,0,0,….
DEF, no,0,1,0,0,1,2
FGH,yes,0,0,0,1,2,3
.
.
.
Till 60
Can u please suggest me a suitable feature selection for my data?
Hi Mohit,
Consider trying a few different methods, as well as some projection methods and see which “views” of your data result in more accurate predictive models.
Hell!
Once I got the reduced version of my data as a result of using PCA, how can I feed to my classifier?
example: the original data is of size 100 row by 5000 columns
if I reduce 200 features I will get 100 by 200 dimension data. right?
then I create arrays of
a=array[:,0:199]
b=array[:,99]
but when I test my classifier its core is 0% in both test and training accuracy?
An7y Idea
Sounds like you’re on the right, but a zero accuracy is a red flag.
Did you accidently include the class output variable in the data when doing the PCA? It should be excluded.
Hello sir,
I have a question in my mind
each of these feature selection algo uses some predefined number like 3 in case of PCA.So how we come to know that my data set cantain only 3 or any predefined number of features.it does not automatically select no features its own.
Great question Kamal.
No, you must select the number of features. I would recommend using a sensitivity analysis and try a number of different features and see which results in the best performing model.
Hi jason,
I have a question about the RFECV approach.
I’m dealing with a project where I have to use different estimators (regression models). is it correct use RFECV with these models? or is it enough to use only one of them? Once I have selected the best features, could I use them for each regression model?
To better explain:
– I have used RFECV on whole dataset in combination with one of the following regression models [LinearRegression, Ridge, Lasso]
– Then I have compared the r2 and I have chosen the better model, so I have used its features selected in order to do others things.
– pratically, I use the same ‘best’ features in each regression model.
Sorry for my bad english.
Good question.
You can embed different models in RFE and see if the results tell the same or different stories in terms of what features to pick.
You can build a model from each set of features and combine the predictions.
You can pick one set of features and build one or models from them.
My advice is to try everything you can think of and see what gives the best results on your validation dataset.
Thank you man. You’re great.
You’re welcome.
Hi Jason.
Thanks for the post, but I think going with Random Forests straight away will not work if you have correlated features.
Check this paper:
https://academic.oup.com/bioinformatics/article/27/14/1986/194387/Classification-with-correlated-features
I am not sure about the other methods, but feature correlation is an issue that needs to be addressed before assessing feature importance.
Makes sense, thanks for the note and the reference.
Jason, following this notes, do you have any ‘rule of thumb’ when correlation among the input vectors become problematic in the machine learning universe? after all, the features reduction technics which embedded in some algos (like the weights optimization with gradient descent) supply some answer to the correlations issue.
Thanks
Perhaps a correlation above 0.5. Perform a sensitivity analysis with different values, select features and use resulting model skill to help guide you.
Hello sir,
Thank you for the informative post. My questions are
1) How do you handle NaN in a dataset for feature selection purposes.
2) I am getting an error with RFE(model, 3) It is telling me i supplied 2 arguments
instead of 1.
Thank you very much once again.
Hi, NaN is a mark of missing data.
Here are some ways to handle missing data:
https://machinelearningmastery.com/handle-missing-data-python/
I solved my problem sir. I named the function RFE in my main but. I would love to hear
your response to first question.
how to load the nested JSON into the data frame ?
I don’t know off hand, perhaps post to StackOverflow Sam?
good afternoon
How to know with pca what are the main components?
PCA will calculate and return the principal components.
Yes but pca does not tell me which are the most relevant varials if mass test etc?
Not sure I follow you sorry.
You could apply a feature selection or feature importance method to the PCA results if you wanted. It might be overkill though.
In RFE we should input a estimator, so before I do feature selection, should I fine tune the model or just use the default parmater settting? Thanks.
You can, but that is not really required. As long as the estimator is reasonably skillful on the problem, the selected features will be valuable.
I was suck here for days. Thanks a lot.
stuck…
I’m glad to hear the advice helped.
I’m here to help if you get stuck again, just post your questions.
Hi Jason,
I was wondering if I could build/train another model (say SVM with RBF kernel) using the features from SVM-RFE (wherein the kernel used is a linear kernel).
Sure.
Hi Jason,
First of all thank you for all your posts ! It’s very helpful for machine learning beginners like me.
I’m working on a personal project of prediction in 1vs1 sports. My neural network (MLP) have an accuracy of 65% (not awesome but it’s a good start). I have 28 features and I think that some affect my predictions. So I applied two algorithms mentionned in your post :
– Recursive Feature Elimination,
– Feature Importance.
But I have some contradictions. For exemple with RFE I determined 20 features to select but the feature the most important in Feature Importance is not selected in RFE. How can we explain that ?
In addition to that in Feature Importance all features are between 0,03 and 0,06… Is that mean that all features are not correlated with my ouput ?
Thanks again for your help !
Hi Gwen,
Different feature selection methods will select different features. This is to be expected.
Build a model on each set of features and compare the performance of each.
Consider ensembling the models together to see if performance can be lifted.
A great area to consider to get more features is to use a rating system and use rating as a highly predictive input variable (e.g. chess rating systems can be used directly).
Let me know how you go.
Thanks for your answer Jason.
I tried with 20 features selected by Recursive Feature Elimination but my accuracy is about 60%…
In addition to that the Elo Rating system (used in chess) is one of my features. With this feature only my accuracy is ~65%.
Maybe a MLP is not a good idea for my project. I have to think about my NN configuration I only have one hidden layer.
And maybe we cannot have more than 65/70% for a prediction of tennis matches.
(Not enough for a positive ROI !)
Hang in there Gwen.
Try lots of models and lots of config for models.
See what skill other people get on the same or similar problems to get a feel for what is possible.
Brainstorm for days about features and other data you could use.
See this post:
https://machinelearningmastery.com/machine-learning-performance-improvement-cheat-sheet/
Hello Jason,
I am very much impressied by this tutorial. I am just a beginner. I have a very basic question. Once I got the reduced version of my data as a result of using PCA, how can I feed to my classifier? I mean to say how to feed the output of PCA to build the classifier?
Assign it to a variable or save it to file then use the data like a normal input dataset.
Hi Jason,
I was trying to execute the PCA but, I got the error at this point of the code
print(“Explained Variance: %s”) % fit.explained_variance_ratio_
It’s a type error: unsupported operand type(s) for %: ‘non type’ and ‘float’
Please help me.
Looks like a Python 3 issue. Move the “)” to the end of the line:
Thanks Jason. It works.
Glad to hear it.
How to know wich feature selection technique i have to choose?
Consider using a few, create models for each and select the one that results in the best performing model.
Hello Jason,
I have used the extra tree classifier for the feature selection then output is importance score for each attribute. But then I want to provide these important attributes to the training model to build the classifier. I am not able to provide only these important features as input to build the model.
I would be greatful to you if you help me in this case.
The importance scores are for you. You can use them to help decide which features to use as inputs to your model.
Hi Jason, I truely appreciate your post. But I have a quick question. Why the sum of the importance scores is unequal to 1?
Because they are not normalized.
I am sincerely grateful to you. I ran feature importance using SelectFromModel with estimator=LinearSVC. But I got negative feature importance values. I would like to kown what that means.
Scores are often relative. Perhaps those features are less important than others?
Thank you very much.
Hi Jason,
Basically i want to provide feature reduction output to Naive Bays. I f you could provide sample code will be better.
Thanks for providing this wonderful tutorial.
You can use feature selection or feature importance to “suggest” which features to use, then develop a model with those features.
Thanks Jason,
But after knowing the important features, I am not able to build a model from them. I don’t know how to giveonly those featuesIimportant) as input to the model. I mean to say X_train parameter will have all the features as input.
Thanks in advance….
A feature selection method will tell you which features you could use. Use your favorite programming language to make a new data file with just those columns.
thanks a lot Jason. You are doing a great job.
Thanks.
I have my own dataset on the Desktop, not the standard dataset that all machine learning have in their depositories (e.g. iris , diabetes).
I have a simple csv file and I want to load it so that I can use scikit-learn properly.
I need a very simple and easy way to do so.
Waiting for the reply.
Try this tutorial:
https://machinelearningmastery.com/load-machine-learning-data-python/
Thanks for this post, it’s very helpful,
What would make me choose one technique and not the others?
The results of each of these techniques correlates with the result of others?, I mean, makes sense to use more than one to verify the feature selection?.
thanks!
Choose a technique based on the results of a model trained on the selected features.
In predictive modeling we are concerned with increasing the skill of predictions and decreasing model complexity.
Sounds that I’d need to cross-validate each technique… interesting, I know that heavily depends on the data but I’m trying to figure out an heuristic to choose the right one, thanks!.
Applied machine learning is empirical. You cannot pick the “best” methods analytically.
Hi Jason,
In your examples, you write:
array = dataframe.values
X = array[:,0:8]
Y = array[:,8]
In my dataset, there are 45 features. When i write like this:
X = array[:,0:44]
Y = array[:,44]
I get some errors:
Y = array[:,44]
IndexError: index 45 is out of bounds for axis 1 with size 0
If you help me, i ll be grateful!
Thanks in advance.
Confirm that you have loaded your data correctly, print the shape and some rows.
1.. What kind of predictors can be used with Lasso?
2. If categorical predictors can be used, should they be re-coded to have numerical values? ex: yes/no values re-coded to be 1/0
3. Can categorical variables such as location (U(urban)/R(rural)) be used without any conversion/re-coding?
Regression, e.g. predicting a real value.
Categorical inputs must be encoded as integers or one hot encoded (dummy variables).
Hi Jason
I am new to ML and am doing a project in Python, at some point it is to recognize correlated features , I wonder what will be the next step? what to do with correlated features? should we change them to something new? a combination maybe? how does it affect our modeling and prediction? appreciated if you direct me into some resources to study and find it out.
best
It is common to identify and remove the correlated input variables.
Try it and see if it lifts skill on your model.
Hello Dr Brownlee,
Thank you for these incredible tutorials.
I am trying to classify some text data collected from online comments and would like to know if there is any way in which the constants in the various algorithms can be determined automatically. For example, in SelectKBest, k=3, in RFE you chose 3, in PCA, 3 again whilst in Feature Importance it is left open for selection that would still need some threshold value.
Is there a way like a rule of thumb or an algorithm to automatically decide the “best of the best”? Say, I use n-grams; if I use trigrams on a 1000 instance data set, the number of features explodes. How can I set SelectKBest to an “x” number automatically according to the best? Thank you.
No, hyperparameters cannot be set analytically. You must use experimentation to discover the best configuration for your specific problem.
You can use heuristics or copy values, but really the best approach is experimentation with a robust test harness.
It was an impressive tutorial, quite easy to understand. I am looking for feature subset selection using gaussian mixture clustering model in python. Can you help me out?
Sorry, I don’t have material on mixture models or clustering. I cannot help.
Hi jason
I’ve tried all feature selection techniques which one is opt for training the data for the predictive modelling …?
Try many for your dataset and see which subset of features results in the most skillful model.
Hello Jason,
I am a biochemistry student in Spain and I am on a project about predictive biomarkers in cancer. The bioinformatic method I am using is very simple but we are trying to predict metastasis with some protein data. In our research, we want to determine the best biomarker and the worst, but also the synergic effect that would have the use of two biomarkers. That is my problem: I don’t know how to calculate which are the two best predictors.
This is what I have done for the best and worst predictors:
analisis=[‘il10meta’]
X = data[analisis].values
#response variable
response=’evol’
y = data[response].values
# use train/test split with different random_state values
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=5)
from sklearn.neighbors import KNeighborsClassifier
#creating the classfier
knn = KNeighborsClassifier(n_neighbors=1)
#fitting the classifier
knn.fit(X_train, y_train)
#predicting response variables corresponding to test data
y_pred = knn.predict(X_test)
#calculating classification accuracy
print(metrics.accuracy_score(y_test, y_pred))
I have calculate the accuracy. But when I try to do the same for both biomarkers I get the same result in all the combinations of my 6 biomarkers.
Could you help me? Any tip?
THANK YOU
Generally, I would recommend following this process to get the best model for your predictive modeling problem:
https://machinelearningmastery.com/start-here/#process
Generally, you must test many different models and many different framings of the problem to see what works best.
Hello Jason,
Many thanks for your post. I have also read your introduction article about feature selection. Which method is Feature Importance categorized under? i.e wrapper or embedded ?
Thanks
Neither, it is a different thing yet again.
You could use the importance scores as a filter.
Great post! Thank you, Jason. My question is all these in the post here are integers. That is needed for all algorithms. What if I have categorical data? How can I know which feature is more important for the model if there are categorical features? Is there a method/way to calculate it before one-hot encoding(get_dummies) or how to calculate after one-hot encoding if the model is not tree-based?
Good question, I cannot think of feature selection methods specific to categorical data off hand, they may be out there. Some homework would be required (e.g. google scholar search).
hello Jason,
Should I do Feature Selection on my validation dataset also? Or just do feature selection on my training set alone and then do the validation using the validation set?
Use the train dataset to choose features. Then, only choose those features on test/validation and any other dataset used by the model.
hello jason
i am doing simple classification but there is coming an issue
ValueError Traceback (most recent call last)
in ()
—-> 1 fit = test.fit(X, Y)
~\Anaconda3\lib\site-packages\sklearn\feature_selection\univariate_selection.py in fit(self, X, y)
339 Returns self.
340 “””
–> 341 X, y = check_X_y(X, y, [‘csr’, ‘csc’], multi_output=True)
342
343 if not callable(self.score_func):
~\Anaconda3\lib\site-packages\sklearn\utils\validation.py in check_X_y(X, y, accept_sparse, dtype, order, copy, force_all_finite, ensure_2d, allow_nd, multi_output, ensure_min_samples, ensure_min_features, y_numeric, warn_on_dtype, estimator)
571 X = check_array(X, accept_sparse, dtype, order, copy, force_all_finite,
572 ensure_2d, allow_nd, ensure_min_samples,
–> 573 ensure_min_features, warn_on_dtype, estimator)
574 if multi_output:
575 y = check_array(y, ‘csr’, force_all_finite=True, ensure_2d=False,
~\Anaconda3\lib\site-packages\sklearn\utils\validation.py in check_array(array, accept_sparse, dtype, order, copy, force_all_finite, ensure_2d, allow_nd, ensure_min_samples, ensure_min_features, warn_on_dtype, estimator)
431 force_all_finite)
432 else:
–> 433 array = np.array(array, dtype=dtype, order=order, copy=copy)
434
435 if ensure_2d:
ValueError: could not convert string to float: ‘no’
can you guide me in this regard
You may want to use a label encoder and a one hot encoder to convert string data to numbers.
import numpy as np
from pandas import read_csv
from sklearn.feature_selection import RFE
from sklearn.linear_model import LogisticRegression
# load data
data = read_csv(‘C:\\Users\\abc\\Downloads\\xyz\\api.csv’,names = [‘org.apache.http.impl.client.DefaultHttpClient.execute’,’org.apache.http.impl.client.DefaultHttpClient.’,’java.net.URLConnection.getInputStream’,’java.net.URLConnection.connect’,’java.net.URL.openStream’,’java.net.URL.openConnection’,’java.net.URL.getContent’,’java.net.Socket.’,’java.net.ServerSocket.bind’,’java.net.ServerSocket.’,’java.net.HttpURLConnection.connect’,’java.net.DatagramSocket.’,’android.widget.VideoView.stopPlayback’,’android.widget.VideoView.start’,’android.widget.VideoView.setVideoURI’,’android.widget.VideoView.setVideoPath’,’android.widget.VideoView.pause’,’android.text.format.DateUtils.formatDateTime’,’android.text.format.DateFormat.getTimeFormat’,’android.text.format.DateFormat.getDateFormat’,’android.telephony.TelephonyManager.listen’,’android.telephony.TelephonyManager.getSubscriberId’,’android.telephony.TelephonyManager.getSimSerialNumber’,’android.telephony.TelephonyManager.getSimOperator’,’android.telephony.TelephonyManager.getLine1Number’,’android.telephony.SmsManager.sendTextMessage’,’android.speech.tts.TextToSpeech.’,’android.provider.Settings$System.getString’,’android.provider.Settings$System.getInt’,’android.provider.Settings$System.getConfiguration’,’android.provider.Settings$Secure.getString’,’android.provider.Settings$Secure.getInt’,’android.os.Vibrator.vibrate’,’android.os.Vibrator.cancel’,’android.os.PowerManager$WakeLock.release’,’android.os.PowerManager$WakeLock.acquire’,’android.net.wifi.WifiManager.setWifiEnabled’,’android.net.wifi.WifiManager.isWifiEnabled’,’android.net.wifi.WifiManager.getWifiState’,’android.net.wifi.WifiManager.getScanResults’,’android.net.wifi.WifiManager.getConnectionInfo’,’android.media.RingtoneManager.getRingtone’,’android.media.Ringtone.play’,’android.media.MediaRecorder.setAudioSource’,’android.media.MediaPlayer.stop’,’android.media.MediaPlayer.start’,’android.media.MediaPlayer.setDataSource’,’android.media.MediaPlayer.reset’,’android.media.MediaPlayer.release’,’android.media.MediaPlayer.prepare’,’android.media.MediaPlayer.pause’,’android.media.MediaPlayer.create’,’android.media.AudioRecord.’,’android.location.LocationManager.requestLocationUpdates’,’android.location.LocationManager.removeUpdates’,’android.location.LocationManager.getProviders’,’android.location.LocationManager.getLastKnownLocation’,’android.location.LocationManager.getBestProvider’,’android.hardware.Camera.open’,’android.bluetooth.BluetoothAdapter.getAddress’,’android.bluetooth.BluetoothAdapter.enable’,’android.bluetooth.BluetoothAdapter.disable’,’android.app.WallpaperManager.setBitmap’,’android.app.KeyguardManage$KeyguardLock.reenableKeyguar’,’android.app.KeyguardManager$KeyguardLock.disableKeyguard’,’android.app.ActivityManager.killBackgroundProcesses’,’android.app.ActivityManager.getRunningTasks’,’android.app.ActivityManager.getRecentTasks’,’android.accounts.AccountManager.getAccountsByType’,’android.accounts.AccountManager.getAccounts’,’Class’])
dataframe = read_csv(url, names=names)
array = dataframe.values
X = array[:,0:70]
Y = array[:,70]
# feature extraction
model = LogisticRegression()
rfe = RFE(model, 3)
fit = rfe.fit(X, Y)
#print(“Num Features: %d”) % fit.n_features_
#print(“Selected Features: %s”) % fit.support_
#print(“Feature Ranking: %s”) % fit.ranking_
—————————————————————————————————————–
I get following error
ValueError Traceback (most recent call last)
in ()
6 model = LogisticRegression()
7 rfe = RFE(model, 3)
—-> 8 fit = rfe.fit(X, Y)
9 print(“Num Features: %d”) % fit.n_features_
10 print(“Selected Features: %s”) % fit.support_
Perhaps try posting your code to stackoverflow?
Can you post a code on first select relevant features using any feature selection method, and then use relevant features to construct classification model?
Thanks for the suggestion.
will you post a code on selecting relevant features using feature selection method and then using relevant features constructing a classification model??
Yes, see this post:
https://machinelearningmastery.com/feature-selection-in-python-with-scikit-learn/
Hi Jason,
Thanks for the content, it was really helpful.
Can you clarify if the above mentioned methods can also be used for regression models?
Perhaps, I’m no sure off hand. Try and let me know how you go.
Hi Jason,
I just had the same question as Arjun, I tried with a regression problem but neither of the approaches were able to do it.
What was the problem exactly?
Hi Jason! Can you please further explain what the vector does in the separateByClass method?
Sorry, I don’t follow?
Hi Jason,
Thank you for the post, it was very useful.
I have a regression problem with one output variable y (0<=y<=100) and 6 input features (I think that they are non-correlated).
The number of observations (samples) is 36980.
I used Random Forest algorithm to fit the prediction model.
The mean absolute error obtained is about 7.
Do you advise me to make features selection or not in this case?
In other words, from which number of features, it is advised to make features selection?
Congratulations.
Try a suite of methods, build models from the selected features and see if the models outperform those models that use all features.
Hello Jason,
First thanks for sharing.
I have question with regards to four automatic feature selectors and feature magnitude. I noticed you used the same dataset. Pima dataset with exception of feature named “pedi” all features are of comparable magnitude.
Do you need to do any kind of scaling if the feature’s magnitude was of several orders relative to each other? For example if we assume one feature let’s say “tam” had magnitude of 656,000 and another feature named “test” had values in range of 100s. Will this affect which automatic selector you choose or do you need to do any additional pre-processing?
The scale of features can impact feature selection methods, it really depends on the method.
If you’re in doubt, consider normalizing the data before hand.
Feature scaling should be included in the examples.
The Pima Indians onset of diabetes dataset contains features with a large mismatches in scale. The rankings produced by the code in this article are influenced by this, and thus are not accurate.
Thanks for the suggestion Eric.
Hello Jason,
One more question:
I noticed that when you use three feature selectors: Univariate Selection, Feature Importance and RFE you get different result for three important features.
1. When using Univariate with k=3 chisquare you get
plas, test, and age as three important features. (glucose tolerance test, insulin test, age)
2. When using Feature Importance using ExtraTreesClassifier
The score suggests the three important features are plas, mass, and age. Glucose tolerance test, weight(bmi), and age)
3. When you use RFE
RFE chose the top 3 features as preg, mass, and pedi. Number of pregnancy, weight(bmi), and Diabetes pedigree test.
According your article below
https://machinelearningmastery.com/an-introduction-to-feature-selection/
Univariate is filter method and I believe the RFE and Feature Importance are both wrapper methods.
All three selector have listed three important features. We can say the filter method is just for filtering a large set of features and not the most reliable? However, the two other methods don’t have same top three features? Are some methods more reliable than others? Or does this come down to domain knowledge?
Different methods will take a different “view” of the data.
There is no “best” view. My advice is to try building models from different views of the data and see which results in better skill. Even consider creating an ensemble of models created from different views of the data together.
Hi Jason,
I’m your fan. Your articles are great. Two questions on the topic of feature selection
1. Shouldn’t you convert your categorical features to “categorical” first?
2. Don’t we have to normalize numeric features
Before doing PCA or feature selection? In my case it is taking the feature with the max value as important feature.
And, not all methods produce the same result.
Any thoughts?
Cheers,
Ranbeer
Yes, Python requires all features to be numerical. Sometimes it can benefit the model if we rescale the input data.
hi jason sir,
your articles are very helpful.
i have a confusion regarding gridserachcv()
i am working on sentiment analyis and i have created different group of features from dataset.
i am using linear SVC and want to do grid search for finding hyperparameter C value. After getting value of C, fir the model on train data and then test on test data. But i also want to check model performnce with different group of features one by one so do i need to do gridserach again and again for each feature group?
Perhaps, it really depends how sensitive the model is to your data.
Also, so much grid searching may lead to some overfitting, be careful.
Thank you Jason for gentle explanation.
The last part “# Feature Importance with Extra Trees Classifier”.
It looks the result is different if we consider the higher scores?
Sorry, what do you mean exactly?
Hi
Sir why you use just 8 example and your dataset contain many example ??
Sorry, I don’t follow. Perhaps you can try rephrasing your question?
Hi Jason,
Your articles are awesome . After going through this article, this is stuck in my mind.
Out of these 4 suggested techniques, which one I have to select ?
Why the O/P is different based on different feature selection?
Thanks
Try them all and see which results in a model with the most skill.
Dear Jason,
Thank you the article.
When I am trying to use Feature Importance I am encountering the following error.
Can you please help me with this.
File “C:/Users/bhanu/PycharmProjects/untitled3/test_cluster1.py”, line 14, in
model.fit(X, Y)
File “C:\Users\bhanu\PycharmProjects\untitled3\venv\lib\site-packages\sklearn\ensemble\forest.py”, line 247, in fit
X = check_array(X, accept_sparse=”csc”, dtype=DTYPE)
File “C:\Users\bhanu\PycharmProjects\untitled3\venv\lib\site-packages\sklearn\utils\validation.py”, line 433, in check_array
array = np.array(array, dtype=dtype, order=order, copy=copy)
ValueError: could not convert string to float: ‘neptune’
Perhaps you are running on a different dataset? Where did ‘neptune’ come from?
Can I get more information about Univariate Feature selection??? I mean more models like ReliefF, correlation etc.,
Thanks for the suggestion.
Hi Jason,
Thank you for the post, it was very useful.
I have a problem that is one-class classification and I would like to select features from the dataset, however, I see that the methods that are implemented need to specify the target but I do not have the target since the class of the training dataset is the same for all samples.
Where can I found some methods for feature selection for one-class classification?
Thanks!
If the class is all the same, surely you don’t need to predict it?
Well, my dataset is related to anomaly detection. So the training set contains only the objects of one class (normal conditions) and in the test set, the file combines samples that are under normal conditions and data from anomaly conditions.
What I understand is that in feature selection techniques, the label information is frequently used for guiding the search for a good feature subset, but in one-class classification problems, all training data belong to only one class.
For that reason, I was looking for feature selection implementations for one-class classification.
Thank you for the post, it was very useful for beginner.
I have a problem that is I use Feature Importance with Extra Trees Classifier and how can
I display feature name(plas,age,mass,….etc) in this sample.
for example:
Feature ranking:
1. plas (0.11070069)
2. age (0.2213717)
3. mass(0.08824115)
…….
Thanks for your help.
……..
You might have to write some custom code I think.
Use the following:
print(list(zip(names, model.feature_importances_)))
You get:
[(‘preg’, 0.11289758476179099), (‘plas’, 0.23098096297414206), (‘pres’, 0.09989914623444449), (‘skin’, 0.08008405837625963), (‘test’, 0.07442780491152233), (‘mass’, 0.14140399156908312), (‘pedi’, 0.11808706393665534), (‘age’, 0.142219387236102)]
Nice!
Hi Jason,
I tried Feature Importance method, but all the values of variables are above 0.05, so does it mean that all the variables have little relation with the predicted value?
Perhaps try other feature selection methods, build models from each set of features and double down on those views of the features that result in the models with the best skill.
Hello Jason,
Thanks for you great post, I have a question in feature reduction using Principal Component Analysis (PCA), ISOMAP or any other Dimensionality Reduction technique how will we be sure about the number of features/dimensions is best for our classification algorithm in case of numerical data.
Try multiple configurations, build and evaluate a model for each, use the one that results in the best model skill score.
Hi,
what to do when i have multiple categorical features like zipcode,class etc
should i hot encode them
Some like zip code you could use a word embedding.
Others like class can be one hot encoded.
Hi,
Iwhen we use univariate filter techniques like Pearson correlation, mutul information and so on. Do we need to apply the filter technique on training set not on the whole dataset??
Perhaps just work with the training data.
jason – i’m working on several feature selection algorithms to cull from a list of about a 100 or so input variables for a continuous output variable i’m trying to predict. these are helpful examples, but i’m not sure they apply to my specific regression problem i’m trying to develop some models for…and since i have a regression problem, are there any feature selection methods you could suggest for continuous output variable prediction?
i.e. i have normalized my dataset that has 100+ categorical, ordinal, interval and binary variables to predict a continuous output variable…any suggestions?
thanks in advance!
RFE will work for classification or regression. It’s a good place to start.
Also, correlation of inputs with the output is another excellent starting point.
I read your post, it was very productive.
Can i use linear correlation coefficient between categorical and continuous variable for feature selection.
or please suggest me some other method for this type of dataset (ISCX -2012) in which target class is categorical and all other attributes are continuous.
No.
Perhaps look into feature importance scores.
Jason,
I was wondering whether the parameters of the machine learning tool that is used during the feature selection step are of any importance. Since most websites that I have seen so far just use the default parameter configuration during this phase.
I understand that adding a grid search has the following consequenses:
-It increase the calculation time substantially. (really when using wrapper (recursive feature elimination))
-Hard to determine which produces better results, really when the final model is constructed with a different machine learning tool.
But still, is it worth it to investigate it and use multiple parameter configurations of the feature selection machine learning tool?
My situation:
-A (non-linear) dataset with ~20 features.
-Planning to use XGBooster for the feature selection phase (a paper with a likewise dataset stated that is was sufficient).
-For the construction of the model I was planning to use MLP NN, using a gridsearch to optimize the parameters.
Thanks in advance!
Yes you can tune them.
Generally, I recommend generating many different “views” on the inputs, fit a model to each and compare the performance of the resulting models. Even combine them.
Most likely, there is no one best set of features for your problem. There are many with varying skill/capability. Find a set or ensemble of sets that works best for your needs.
Hi Jason
I need to do feature engineering on rows selection by specifying the best window size and frame size , do you have any example available online?
thanks
Sa
For time series, yes right here:
https://machinelearningmastery.com/sensitivity-analysis-history-size-forecast-skill-arima-python/
Hi Jason
I am a beginner in python and scikit learn. I am currently trying to run a svm algorithm to classify patheitns and healthy controls based on functional connectivity EEG data. I’m using rfcv to select the best features out of approximately 20’000 features. I get 32 selected features and an accuracy of 70%. What I want to try next is to run a permutation statistic to check if my result is significant.
My question: Do I have to run the permutation statistic on the 32 selected features? Or do I have to include the 20’000 for this purpose.
Below you can see my code. to simplify my question, i reduced the code to 5 features, but the rest is identical. I would appreciate your help very much, as I cannot find any post about this topic.
Best Yolanda
homeDir = ‘F:\Analysen\Prediction_TreatmentOutcome\PyCharmProject_TreatmentOutcome’ # location of the connectivity matrices
# #############################################################################
# import packages needed for classification
import numpy as np
import os
import matplotlib.pyplot as plt
from sklearn.datasets import make_classification
from sklearn.model_selection import train_test_split
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import cross_validate
from sklearn.model_selection import StratifiedKFold
from sklearn.feature_selection import RFECV
from sklearn import svm
from sklearn.pipeline import make_pipeline, Pipeline
from sklearn import preprocessing
from sklearn.model_selection import permutation_test_score
class PipelineRFE(Pipeline):
def fit(self, X, y=None, **fit_params):
super(PipelineRFE, self).fit(X, y, **fit_params)
self.coef_ = self.steps[-1][-1].coef_
return self
clf = PipelineRFE(
[
(‘std_scaler’, preprocessing.StandardScaler()), #z-transformation
(“svm”, svm.SVC(kernel = ‘linear’, C = 1)) #estimator
]
)
# #############################################################################
# Load and prepare data set
nQuest = 5 # number of questionnaires
samples = np.loadtxt(‘FBDaten_T1.txt’)
# Import targets (created in R based on group variable)
targets = np.genfromtxt(r’F:\Analysen\Prediction_TreatmentOutcome\PyCharmProject_TreatmentOutcome\Targets_CL_fbDaten.txt’, dtype= str)
# #############################################################################
# run classification
skf = StratifiedKFold(n_splits = 5) # The folds are made by preserving the percentage of samples for each class.
# rfecv
rfecv = RFECV(estimator = clf, step = 1, cv = skf, scoring = ‘accuracy’)
rfecv.fit(samples, targets)
# The number of selected features with cross-validation.
print(“Optimal number of features : %d” % rfecv.n_features_)
# Plot number of features VS. cross-validation scores
plt.figure()
plt.xlabel(“Subset of features”)
plt.ylabel(“Cross validation score (nb of correct classifications)”)
plt.plot(range(1, len(rfecv.grid_scores_) + 1), rfecv.grid_scores_)
plt.show()
#The mask of selected features
rfecv.support_
print(“Mask of selected features : %s” % rfecv.support_)
#Find index of true values in a boolean vector
index_features = np.where(rfecv.support_)[0]
print(index_features)
#Find value of indices
reduced_features = samples[:, index_features]
print(reduced_features)
## permutation testing on reduced features
score, permutation_scores, pvalue = permutation_test_score(
clf, reduced_features, targets, scoring=”accuracy”, cv=skf, n_permutations=100, n_jobs=1)
print(“Classification score %s (pvalue : %s)” % (score, pvalue))
Sorry, I do not have the capacity to review your code.
Thank you a lot for this useful tutorial. It would’ve been appreciated if you could elaborate on the cons/pros of each method.
Thanks in advance.
Thanks for the suggestion.
I want to ask about feature extraction procedure, what’s the criteria to stop training and extract features. Are it depend on the test accuracy of model?. In other meaning what is the difference between extract feature after train one epoch or train 100 epoch? what is best features?, may be my question foolish but i need answer for it.
What do you mean by extract features? Here, we are talking about feature selection?
I ask about feature extraction procedure, for example if i train CNN, after which number of epochs should stop training and extract features?. In other meaning are feature extraction depend on the test accuracy of training model?. If i build model (any deep learning method) to only extract features can i run it for one epoch and extract features?
I see.
You want to use features from a model that is skillful. Perhaps at the same task, perhaps at a reconstruction task (e.g. an autoencoder).
I do not understand answer
Sorry, which part?
Hello Sir,
Thank you soo much for this great work.
Will you please explain how the highest scores are for : plas, test, mass and age in Univariate Selection. I am not getting your point.
What problem are you having exactly?
Thank you sir for the reply…
Actually I was not able to understand the output of chi^2 for feature selection. The problem has been solved now.
Thanks a lot.
I’m happy to hear that you solved your problem.
Which is the best technique for feature selection? and i want to know why the ranking is always change when i try multiple times?
This is a common question that I answer here:
https://machinelearningmastery.com/faq/single-faq/what-feature-selection-method-should-i-use
Hi,
in your example for feature importance you use as Ensemble classifier the ExtraTreesClassifier.
In sci-kit learn the default value for bootstrap sample is false.
Doesn’t this contradict to find the feature importance? e.g it could build the tree on only one feature and so the importance would be high but does not represent the whole dataset.
Thanks
Paul
Trees will sample features and in aggregate the most used features will be “important”.
It only means the features are important to building trees, you can interpret it how ever you like.
Hi Jason,
I have a dataset which contains both categorical and numerical features. Should I do feature selection before one-hot encoding of categorical features or after that ?
Sure. It’s a cheap operation (easy) and has big effects on performance.
Hi Jason
I haven’t read all the comments, so I don’t know if this was mentioned by someone else. I stumbled across this:
https://hub.packtpub.com/4-ways-implement-feature-selection-python-machine-learning/
It’s identical (barring edits, perhaps) to your post here, and being marketed as a section in a book. I thought I should let you know.
That is very disappointing!
Thanks for letting me know Dean.
Hi Jason
I have a dataset with two classes. In the feature selection, I want to specify important features for each class. For example, if I chose 15 important features, determine which attribute is more important for which class.please help me
Yes, this is what feature selection will do for you.
Hi Jason
First, congratulations on your posts and your books.
I am reaing your book machine learning mastery with python and chapter 8 is about this topic and I have a doubt, should I use thoses technical with crude data or should I normalize data first? I did test both case but results are different, exemple (first case column A and B are important but second case column C and D are important)
Very thanks.
Thanks.
Build models from each and go with the approach that results in a model with better performance on a hold out dataset.
HI Jason,
I’m on a project to predict next movement of animals using their past data like location, date and time. what are the possible models that i can use to predict their next location ?
I recommend following this process for new problems:
https://machinelearningmastery.com/start-here/#process
hi,
Many thanks for your hard work on explaining ML to the masses.
I’m trying to optimize my Kaggle-kernel at the moment and I would like to use feature selection. Because my source data contains NaN, I’m forced to use an imputer before the feature selection.
Unfortunately, that results in actually worse MAE then without feature selection.
Do you have a tip how to implement a feature selection with NaN in the source data?
Perhaps you can remove the rows with NaNs from the data used to train the feature selector?
Hi Jason,
Somehow ur blog almost always has exactly what I need. Least I could do is say thanks and wish u all the best!
Thanks!
Hi Jason,
Your work is amazing. Got interested in Machine learning after visiting your site. Thank You, Keep up your good work.
I tried using RFE in another dataset in which I converted all categorical values to numerical values using Label Encoder but still I get the following error:
—————————————————————————
ValueError Traceback (most recent call last)
in ()
14 model = LogisticRegression()
15 rfe = RFE(model, 5)
—> 16 fit = rfe.fit(X, Y)
17 print(“Num Features: %d” % fit.n_features_)
18 print(“Selected Features: %s” % fit.support_)
~\Anaconda3\lib\site-packages\sklearn\feature_selection\rfe.py in fit(self, X, y)
132 The target values.
133 “””
–> 134 return self._fit(X, y)
135
136 def _fit(self, X, y, step_score=None):
~\Anaconda3\lib\site-packages\sklearn\feature_selection\rfe.py in _fit(self, X, y, step_score)
140 # self.scores_ will not be calculated when calling _fit through fit
141
–> 142 X, y = check_X_y(X, y, “csc”)
143 # Initialization
144 n_features = X.shape[1]
~\Anaconda3\lib\site-packages\sklearn\utils\validation.py in check_X_y(X, y, accept_sparse, dtype, order, copy, force_all_finite, ensure_2d, allow_nd, multi_output, ensure_min_samples, ensure_min_features, y_numeric, warn_on_dtype, estimator)
571 X = check_array(X, accept_sparse, dtype, order, copy, force_all_finite,
572 ensure_2d, allow_nd, ensure_min_samples,
–> 573 ensure_min_features, warn_on_dtype, estimator)
574 if multi_output:
575 y = check_array(y, ‘csr’, force_all_finite=True, ensure_2d=False,
~\Anaconda3\lib\site-packages\sklearn\utils\validation.py in check_array(array, accept_sparse, dtype, order, copy, force_all_finite, ensure_2d, allow_nd, ensure_min_samples, ensure_min_features, warn_on_dtype, estimator)
431 force_all_finite)
432 else:
–> 433 array = np.array(array, dtype=dtype, order=order, copy=copy)
434
435 if ensure_2d:
ValueError: could not convert string to float: ‘StudentAbsenceDays’
I am in dire need of a solution for this. Kindly help me .
It suggests your data file may still have string values.
Perhaps double check your loaded data?
I had checked the data type of that particular column and it is of type int64 as given below:
In:
mod_StudentData[‘StudentAbsenceDays’].dtype
Out[]:
dtype(‘int64’)
Nice work!
Hi Jason…
Great article as usual.
I’m novice in ML and the article leaves me with a doubt.
The SelectKBest, RFE and ExtraTreesClassifier are performing Feature Extraction and PCA is performing Feature Extraction.
Am I right with this?
Thanks Jason
Yes.
Hi Jason,
First of all nice tutorial!
My question is that I have a samples of around 30,000 with around 150 features each for a binary classification problem. The plan is to do RFE on GRID SEARCH (Select features and tune parameters at the same pipeline) using a 3-fold cross validation (Each fold, the data is split twice, one for RFE and another for GRID SEARCH), this is done on the entire data set.
Now, after determining the best features and parameters, using the SAME data set, I split it into training / validation / test set and train a model using the selected features and parameters to obtain its accuracy (of the best model possible, and on the test set, of course).
Is this the correct thing to do? My reason for this methodology is that, the feature/parameter selection is a whole different process from the actual model fitting (using the selected features and parameters), meaning the actual model fitting will not actually know what the feature/parameter selection learned on the entire dataset, hence it is only okay to re-use the entire data set.
If this is not the case, what would you recommend? perhaps, separate the entire data set into a feature/parameter selection set and actual model fitting set (50:50), wherein after the best features and parameters have been determined on the first 50%, use these features on the remaining 50% of the data to train a model (this 50 is further split into train/validation/test).
I recommend performing feature selection on each fold of CV or with a separate dataset up front.
Thank you for the answer Dr. Jason! Also, the grid selection + RFE process is going to spit out the accuracy / F1-score of the best model attained (with best feature set and parameters), can this be considered as the FINAL score of the your model’s performance? or do you really need to build another model (the final model with your best feature set and parameters) to get the actual score of the model’s performance?
I recommend building a final model for making predictions. The score from the test harness may be a suitable estimate of model performance on unseen data -really it is your call on your project regarding what is satisfactory.
Thanks Dr. Jason. One last question :), can I use the chi squared statistical test (in the univariate selection portion) for reflecting the p-value or the statistical significance of each feature? Let say, I am going to show the trimmed mean of each feature in my data, does the chi squared p-value confirm the statistical significance of the trimmed means?
No, it comments on the relationship between categorical variables.
Thanks Dr. Jason!.. One last question promise 🙂 I assume its okay to prune my features and parameters on a 3-fold nested cross-validated RFE + GS while building my final model on a 10-fold regular cross validation. I used different data sets on each process (I split the original dataset 50:50, used the first half for RFE + GS and the second half to build my final model). The reason is that the nested cross-validated RFE + GS is too computationally expensive and that I’d like to train my final model on a finer granularity hence, the regular 10-fold CV.
Thank you soo much!
I cannot comment if your test methodology is okay, you must evaluate it in terms of stability/variance and use it if you feel the results will be reliable.
Thank you so much Dr. Jason
Hi Dr. Jason;
I want to ask you a question: I want to apply the PSO algorithm in a dataset similar to Pima Indians onset of diabetes dataset, I am disturbed, what can I do
Sorry, I don’t have examples of using global optimization algorithms for feature selection – I’m not convinced that the techniques are relatively effective.
Hi,
I am a beginner and my question may be wrong. can we use these feature selection methods in an autoencoder that our inputs and outputs of our network are an image for example mnist? Thanks
Not really, you would be performing feature selection on pixel values.
The autoencoder is doing a form of this for you.
Hi Jason,
All the techniques mentioned by you works perfectly if there is a target variable (Y or 8th column in your case). The dataset i am working on uses unsupervised learning algorithm and hence does not have any target/dependent variable. Does the feature selection work in such cases? If yes, how should i go about it.
It can, but you may have to use a method that selects features based on a proxy metric, like information or correlation, etc.
I don’t have a worked example, sorry.
Hello Jason,
Your blog and the way how you explain things are fantastic! I have a doubt related to feature selection, for real applications, the fit method of some feature selection techniques must be applied just to the training set or to the whole data set (training + testing)?
Thanks a lot!
It is fit on just the training dataset when evaluating a model. It is fit on all data when developing a final model:
https://machinelearningmastery.com/train-final-machine-learning-model/
Thank you very much!
Hello Jason,
First of all thank you for such an informative article.
I need to perform a feature selection using the Filter, Wrapper and Embedded methods. The plan is to then take an average of scores from each selection procedure and select the top 10 features.
My plan is to split the data initially into train and hold out sets. I plan to then use cross-validation for each of the above 3 methods and use only the train data for this (internally in each of the fold). Once i get my top 10 features , i will then only use them in the hold out set and predict my model performance.
Do you feel this method would give me a stable model ? If not, what can i improve / change ?
Thanks in advance.
Perhaps try it and see.
Hi Jason,
thank you about your efforts,
I want to ask about Feature Extraction with RFE
I use your mention code
names = [‘preg’, ‘plas’, ‘pres’, ‘skin’, ‘test’, ‘mass’, ‘pedi’, ‘age’, ‘class’]
and the results are :
[1 2 3 5 6 1 1 4]
when I change the order of columns names as I mention
names = [‘pedi’,’preg’, ‘plas’, ‘pres’, ‘test’, ‘age’, ‘class’,’mass’,’skin’]
I get same output
[1 2 3 5 6 1 1 4]
can you explain how it work?
thank you
Changing the order of the labels does not change the order of the columns in the dataset. This is why you are getting the same output indexes.
I try to change the order of columns to check the validity of the RFE rank.
what is your advice if I want to check the validity of rank?
Also, I want to ask when I try to choose the features that influence on my models, I should add all features in my dataset ( numerical and categorical) or only categorical features?
Thank you
I’m not sure this is required.
Nevertheless, you would have to change the column order in the data itself, e.g. the numpy array or CSV from which it was loaded.
It depends on the capabilities of the feature selection method as to what features to include during selection.
I am unable to get output, because of this warning:
“C:\Users\Waqar\Anaconda3\lib\site-packages\sklearn\model_selection\_split.py:626: Warning: The least populated class in y has only 1 members, which is too few. The minimum number of members in any class cannot be less than n_splits=5.”
Please help if someone knows.
Perhaps you can use fewer splits or use more data?
Hi Jason,
I am reading from your book on ML Mastery with Python and I was going to the same topic mentioned above, I see you have chose chi square to do feature selection in univariate method, how do I decide to choose between different tests (chi square, t-test , ANOVA).
Great question. I answer it here:
https://machinelearningmastery.com/faq/single-faq/what-feature-selection-method-should-i-use
Thank you, a big post to read for next learning steps 🙂
Thanks.
hi jason
please let me how to choose best k value in case of using SelectKBest this class.
Evaluate models for different values of k and choose the value for k that gives the most skillful model.
hi jason,
i want to use Univariate selection method.
I am building a linear regression model which has around 46 categorical variables.
if i want to know the best categorical features to be used in building my model, I need to send onehot encoded values to fit function right and the score_func sjould also be chi2 ?
test = SelectKBest(score_func=chi2, k=4)
fit = test.fit(X, Y)
In the above code X should be the one hot encoded values of all the categorical variables right ?
Thanks in advance
Perhaps this will help:
https://machinelearningmastery.com/chi-squared-test-for-machine-learning/
Also, linear regression sounds like a bad fit, try a decision tree, and some other algorithms as well.
Hi,
I had a question.
Do you apply feature selection before creating the dummies or after?
Thanks in advance
Feature selection is performed before.
Dear Sir,
I am working on feature selection using “Removing features with low variance”. I am having 1452 features and code is returning me 454 features but with no feature labels i.e column headers. Know I am unable to get that which feature have been accepted. So My question ‘”how I can retain column headers in my output”?
Often feature selection methods choose the column index. Once you have the column index, you can use it on the original data to get the heads for each chosen column.
Hi Jason,
Chi2 test is applicable only for categorical data. But in your example you are using continuous features. I am not sure about it, does SelectKBest is doing any kind of binning to apply Chi2 on continuous data please explain. I also have similar data with continuous features and binary class. I want to be sure before using this method.
Thanks,
Bharat
It is really only used for ordinal/categorical data, e.g. counts and such.
More here:
https://scikit-learn.org/stable/modules/generated/sklearn.feature_selection.chi2.html
Hi Jason,
If i have to figure out which feature selection method is applicable for the kind of data I have, (say) I have to select few features that contributes much for my Target with both Target and Predictor as -Continuous or Categorical or Continuous and Categorical.
Also, Can i just implement one of the techniques that is considered best for all such cases or try few techniques and come to a conclusion.
Thank you, Awaiting your response.
This is a common question that I answer here:
https://machinelearningmastery.com/faq/single-faq/what-feature-selection-method-should-i-use
Can you please list me the best methods or techniques to implement feature selection ..
As mentioned in the link, there is no idea of “best”, instead, you must discover what works well for your specific dataset and choice of model.
Thanks for your reply
No problem.
Hey Jason
Thank you for great tutorial. I just wonder how is the score calculated in chi-squared test? I mean is there any math formula for getting this score? If you know can you explain?
Thank you in advance
Yes, see this post:
https://machinelearningmastery.com/chi-squared-test-for-machine-learning/
Hello Jason,
Thank you for the informative post.
I used GenericUnivariateSelect() from feature_selection.
I have following question regarding this:
1. it says that for mode we have few options to select from i.e: “mode : {‘percentile’, ‘k_best’, ‘fpr’, ‘fdr’, ‘fwe’} Feature selection mode.” Is the K_best of this mode same as SelectKBest function or is it different? If different can you explain to me how this works for scoring and providing the pvalues?
2. What is the significance of pvalues in this output? I understand that usually when we perform statistical test we prefer to select the datapoints with pvalues less that 0.05. So in the output of the selected features if the features have pvalues more than 0.05, is it advisable to drop those features from the list? or the pvalues are not to be considered?
Thank you ! I will be waiting for your response.
Good question, I’m not sure off the cuff. Perhaps check the API documentation?
Hi Jason,
Thanks for your tutorial
I have NaN values in my data and i get the “Input contains NaN, infinity or a value too large for dtype(‘float64’).”
Is there any way to transform data with NaN values. Like how xgboost classifier can work with these values?
Yes, it is a good idea to replace nans with real values before processing, e.g. perhaps with a mean or median value.
Good Afternoon,
When I try the sample code for recursive feature elimination, I receive the following message:
Num Features: %d
Traceback (most recent call last):
File “rfe.py”, line 16, in
print(“Num Features: %d”) % fit.n_features_
TypeError: unsupported operand type(s) for %: ‘NoneType’ and ‘int’
When I run the code for principle component analysis, I get a similar error:
Explained Variance: %s
Traceback (most recent call last):
File “pca.py”, line 16, in
print(“Explained Variance: %s”) % fit.explained_variance_ratio_
TypeError: unsupported operand type(s) for %: ‘NoneType’ and ‘float’
This is after copying and pasting the code exactly and ensuring all whitespace is preserved and that all libraries are up to date. I use the version of python included with my anaconda distro: 3.6. Any assistance would be greatly appreciated as I’m not finding much on stack exchange or anywhere else.
Thank you!
Sorry to hear that, I have some suggestions here that may help:
https://machinelearningmastery.com/faq/single-faq/why-does-the-code-in-the-tutorial-not-work-for-me
Here’s advice on how to run from the command line:
https://machinelearningmastery.com/faq/single-faq/how-do-i-run-a-script-from-the-command-line
How to copy code from the tutorial:
https://machinelearningmastery.com/faq/single-faq/how-do-i-copy-code-from-a-tutorial
Does that help?
Following the suggestion in “Why does the code in the tutorial not work for me,” I went back to StackOverflow and refined my search. (*I mistakenly typed stack exchange, previously.)
Here’s the link for where I found the solution to my problem: https://stackoverflow.com/questions/41788814/typeerror-unsupported-operand-types-for-nonetype-and-float
The code as written here is:
print(“Explained Variance: %s”) % fit.explained_variance_ratio_
All I needed to do to get it to work was:
print((“Explained Variance: %s”) % fit.explained_variance_ratio_)
It’s too simple and I didn’t see it. Thanks for all your help!
Thanks, I have updated the tutorial!
Good article Jason, I run it for my dataset. When I asked for k=1, the best score was 0.01 in the column 101. when I asked for k=4 best, I got
column 58 (score= 0.02)
column 62 (score= 0.001)
column 73 (score= 0.0001 )
column 101(score= 0.01 )
But when I asked for k=2 I got
column 73 (score= 0.0001 )
column 101(score= 0.01 )
It seems SelectKBest already choose the n best and deliver the k best from last column. Am I right? or something else happen??
K-best will select the k best features ordered by the calculated score.
Therefore, should give columns 58 and 101 not 73 and 101.
If you use SelectKBest, it will select the features with the best score for you.
Hey Jason,
Can this is also applicable for Categorical data?
If no, then please suggest other algorithm .
Thanks
Good question.
The chi squared and mutual information functions can be used for feature selection with categorical inputs and class label targets.
Hello,
In one of your post, you mentioned that feature selection methods are:
1. Filter Methods
2. Wrapper Methods
3. Embedded Methods
In this post you say that Feature selection methods are:
univariate selection, feature importance, etc
What is the difference between these methods?
Why are there 2 different posts for the same topic?
The two main types are filter and wrapper, and also perhaps embedding – but that might be a feature engineering method.
Another way to think about it is the number of variables used in the method – univariate or multivariate.
Feature importance is an input to filter methods.
Hi, Jason
Can REFCV only works for sinlge y output data? If I have a 2 outputs regression data, how can I use REFCV to select features?
Thank you
Yes.
Good question. I think something custom is required – perhaps try experimenting.
Hello sir i want to remove all irrelevant features by ranking this feature using Gini impurity index and then select predictors that have non-zero MDI. Please help me
Sorry, I don’t have a tutorial on exactly this.
Hi Jason,
testing RFE feature selection for a logistic regression searching for the best feature, I get different results compared to fitting the model for the individual features and finding the best feature by minimizing the AIC.
e.g.:
RFE finds feature A with:
AIC: 2.4296372383358187 and observed and forecasted: 0 not observed and forecasted: 0 not forecasted and observed: 92 not forecasted and not observed: 1498
minimizing AIC yields feature B with:
AIC 2.337092886023634 and observed and forecasted: 3 not observed and forecasted: 13 not forecasted and observed: 89 not forecasted and not observed: 1485
I assume that RFE uses another score to find the best feature. I thought of applying RFE to identify the best features for different numbers of features [1 to n] and finding the best set by the minimum of the [n] AIC values to circumvent the stepwise regression problem.
But now I am not sure because both steps seem to rely on different scores ?
Many thanks for your help in advance !
Yes, you will get many different perspectives on what good features might be.
Test each and see what results in a model with the best skill for your specific dataset.
hi jason
thnx for your post
i wonder is it better to use feature selection inside cross validation
Yes, it should be used within cross validation to avoid data leakage.
thnx for your reply, but i wonder if you could help me with \that
and i have another question:
in feature importance code
no.of features are 8 and the outputs are 7 how did you know the name of the important features
The example here will help:
https://machinelearningmastery.com/automate-machine-learning-workflows-pipelines-python-scikit-learn/
You can test different cut-off values for importance and discover what works best for your specific dataset.
Hi Jason .
I’m facing this issue .This my code
##########################################################
mtcars_data = pd.read_csv(“D:\Python\Assignment solutions\mtcars.csv”)
# Feature Importance with Extra Trees Classifier
from pandas import read_csv
from sklearn.ensemble import ExtraTreesClassifier
# load my data
array = mtcars_data.values
X = array[:,1:]
Y = array[:,:-10]
# feature extraction
model = ExtraTreesClassifier(n_estimators=10)
model.fit(X, Y)
print(model.feature_importances_)
#####################################################
NotFittedError: Estimator not fitted, call
fitbeforefeature_importances_.That is surprising, sorry to hear that.
I don’t see anything obvious. Perhaps try posting your code and error to stackoverflow?
Thank you so much, your post is very useful to me in knowing the best features to select.
You’re welcome.
Hello Jason, you have shared with us 4 ways to select features, each one of them with diferent answers. How do you select between the 4 approaches? Thanks for such a instructive post.
Good question, this will help:
https://machinelearningmastery.com/faq/single-faq/what-feature-selection-method-should-i-use
Hello Jason,
Nice post, it really helped me.
I am wondering can statistical hypothesis testing be used for feature selection for predictive models, where the target variable is continuous and the predictors are categorical .
Can we use t test, anova, chi-squared test for feature selection?
Yes, see this tutorial:
https://machinelearningmastery.com/feature-selection-with-real-and-categorical-data/
Dear sir,
I go through your blog, regarding recursive feature elimination, please could you help me without using inbuilt method for rfe ( feature selection process.)
Thanks, I have a tutorial on the topic coming.
Thank you sir for your reply. I am looking forward for your tutorial can you please tell me when it will be available.
Yes, get notified here:
https://machinelearningmastery.com/newsletter/
Thank you sir.
You’re welcome.
Dear sir,
I am working with Recursive feature elimination(RFE) using SVM classifier with linear kernel, i have a bit confusion regarding how the internal process of RFE going on, starting it build with all the features ,then how we find the importance of each feature?.How it removing features step by step…can you please explain me in detail ?
I have a tutorial on the method coming.
Until then, perhaps this will help:
https://link.springer.com/article/10.1023%2FA%3A1012487302797
Thank you sir.
You’re welcome.
Hi Sir
I want to apply a muti-layer CNN for classification tasks and the dataset is multi-class and it conatins categorical features . I used to chi-square method for feature selection. Is that is valid point to use chi-square method for feature selection before DNN ?
Thanks
Good question, this will help you choose a feature selection method:
https://machinelearningmastery.com/feature-selection-with-real-and-categorical-data/
The CNN can probably perform a type of feature selection/feature extraction automatically.
Hello Jason,
Thanks a lot for yet another awesome post!
My dataset has over 200 variables and I am running a classification model on it, which is leading to a model OverFit. Which of these methodology do you suggest for reducing the number of features? I started with Feature Importance, however due to such a large number of variables, I am unable to visualise it. Is there a way I can plot or showcase these values with respect to the given variable?
Perhaps use controlled experiments and discover what works best for your dataset.
hi jason,happy to connect with another question.Have the model improved performance after doing this feature extraction? I
Sure. Depends on the dataset and choice of model.
Hello Sir,
Thank you for such a great tutorial on Feature Selection.
The presented methods compare features with a single column (or “variable”?).
How should I perform feature selection, if my features are not of a single column?
For example, if I want to perform classification on an audio dataset, I may extract MFCC Features, RMS Energy, etc for an audio file.
How should I compare MFCC (has 12 cols) and RMS Energy(Single col)?
How should I compare two multi-col features?
Sorry, I don’t have tutorials on working with audio data / DSP.
Actually, I am not asking specifically for audio.
What I am asking is that if the extracted features are comprising of multiple columns themselves, then how do I apply the above methods for feature selection on them?
Feature selection for time series/sequence problems may require specialized methods. Sorry, I don’t have an example.
Hello, thank you for this useful post!
I would like to ask the same thing for Principle Component Analysis and Feature Importance. I have 2686 number of samples and 86 features. Is it possible to keep all the n_samples and just reduce the number of features using this two methods? If yes how is the way to do it?
For example, applying the PCA in my dataset, when I set the n_components=2686, then I have this error:
ValueError: n_components=2686 must be between 0 and min(n_samples, n_features)=86 with svd_solver=’full’
Thank you very much in advance
PCA does not perform feature importance, it creates new features using linear algebra.
Yes, you can use a sequence of feature selection and dimensionality reduction methods in a pipeline if you wish.
Thank you, I will try it.
Best, Despoina
Let me know how you go.
thanks, but if I am want to print the name of selected features, what can I do?
The indexes returned from feature selection can be used as indexes into a list of your column names.
You can see an example in this tutorial:
https://machinelearningmastery.com/rfe-feature-selection-in-python/
hi in wrapper based feature selection why k feature is selected in random way like top 3,top 5 so on .. can anyone answer this??
It is not selected random, we must choose a value that works best for our model and dataset.
Thanks, If I want to selected feature from csv file, what is the code?
Example: file = pd.read_csv(“dataset.csv”)
#select feature
Perhaps see this example:
https://machinelearningmastery.com/rfe-feature-selection-in-python/
Hey Jason, I’m currently working on the project of my school.
We have a dataset of different patients whose vitals are hourly checked. Vitals for example like Blood-Pressure, The PH, Hearth rate. So we have 10 hours of vitals for each patient (with lots of missing data). Then we have to predict, for example, if they will die or not in the following hour.
Our Data per patient looks like this:
Patient Nr Hour Blood-Pressure Hearth Rate PH
1 1 Nan 80 Nan
1 2 Nan 78 Nan
1 3 Nan 75 7.1
1 4 30 Nan Nan
1 5 35 Nan Nan
1 6 30 Nan Nan
1 7 Nan Nan Nan
1 8 Nan 78 7.2
1 9 Nan 75 Nan
1 10 Nan 80 Nan
We substituted the missing values using a SimpleImputer with the mean strategy. We flattened our data so that basically per patient we only have one row of data and tried to fit this with linear regression, but I feel like this approach is like trying to start a fire using two rocks, doable but there surely is a better way. And the resulting predictions are bad.
Do you have any suggestions for the 1)General Approach 2) Strategy to use for filling out missing values 3)Which model to use 4) Maybe a good tutorial that would help me go more into details?
Thank you a lot for this blog
Perhaps you can try modeling the problem as time series classification.
Perhaps some of these suggestions will help:
https://machinelearningmastery.com/faq/single-faq/how-do-i-handle-discontiguous-time-series-data
I’m sorry the initial greeting isn’t very formal, you’re a PhD and I’m a student struggling with my assignment. Hello Doctor Brownlee would be more appropriate
🙂
No, please call me “jason”. This is just a blog and I’m just a guy helping people on the internet.
Dear all
This is a wonderful website and I love it. I’ve got most of my expertise in using python for machine learning and deep learning here, appreciate it very much.
Thanks!
Hi Jason Brownlee,
Thanks for the tutorial
Can you please tell me what should be the value of n_estimators in ExtraTreesClassifier, if I want to select the top ten features from 2048 features?
Keep increasing the value until no further improvement is seen in model performance.
@ Jason Brownlee Thank you for the response.
can you tell me how I will get to know that there is no further improvement in the model performance because by using ExtraTreesClassifier, I will only get the important features that will eventually change with the changing n_estimators.
The only well to tell if there is an improvement with a different configuration is to fit the model with the configuration and evaluate it.
Hello sir
How can i combine mRMR algorithm with CNN or (any pre trained models.
Do you have any idea or any reference code?
Thanks for your efforts…
What is “mRMR”?
mrmr is a feature selection algorithm
Sorry I don’t know anything about that method.
if I want to apply feature selection techniques in deep learning in intrusion detection problem, is it possible?
Why not! Did you try anything? What is the result?
Hello Dr. Jason,
I am trying to use feature selection for classification. I have 36 features in my dataset, I wanted to run multiple feature selection methods to calculate the feature score of all the features. How can I calculate the feature scores of all the features using the feature selection method? Can you please help me?
Good question, see this:
https://machinelearningmastery.com/feature-selection-with-real-and-categorical-data/
My work is to develop a model for multioutput prediction (i.e., predicting five outputs by a single model). When I applied Kbest and recursive feature elimination methods to select the best features, I got an error ‘ bad input shape (x, 5)’.
Does it mean that we can apply these feature selection algorithms only for a single output prediction problem?
No. It just means the model expects an array in one shape while you provided one in a different shape. What you need to do here is to check our input and output and correct it.
Sorry, but I didn’t understand your answer. Maybe I was not able to explain my question.
I have an input array with shape (x,60) and an output array with shape (x,5).
Now when I am applying the SelectKbest algorithm to reduce the number of features of the input vector from 60 to 20, it gives an error “Bad input shape (x,5)”.
However, if I apply an output vector of shape (x,) with only one column, SelectKbest gives the required output.
So it means the output vector should be a 1-D array?
So my question is, how will the SelectKbest work for a multioutput problem where the columns in the output vector can be greater than one?
Ok, that’s right. RFE’s fit(X,y) function expects the y to be a vector, not matrix. Hence you cannot have output of shape (x,5) – this is just a limitation from scikit-learn’s RFE but the theory can still apply if you can define how to measure the error for a 5-dimensional output.
Hello. Thank you very much for sharing such valuable information. I like your content a lot. I have a question: After performing the feature selection, can we already use the most important features in the model or should we check that the most important features are not correlated with each other (from a correlation graph, for example)? Thanks!
Hi Jessica…Yes, I recommend always checking correlation as opposed to making assumptions regarding it. The following may also be of interest to you:
https://machinelearningmastery.com/optimization-for-machine-learning/
Which feature selection method will be suitable if we have a mixed dataset having both numerical as well as categorical data?
Hi Adil…You may find some clarity in the following resource:
https://towardsdatascience.com/learn-how-to-do-feature-selection-the-right-way-61bca8557bef
Thanks for sharing this code. I run the PCA example code, but I think there is an issue because the dimension of the loaded dataset is (768,8) so we have 768 samples and 8 features. By applying PCA we are going to find n_components (here =3) new features so we should have (768,3) after applying PCA. But the written code gives us a dataset with this dimension: (3,8)
.i.e the reduction is applied on samples not on features.
I think a transpose should be applied on X before PCA.
Hi Fatemeh…Did you implement your idea?
Hi James,
I’m a little bit confused with this post and this post “https://machinelearningmastery.com/feature-selection-with-numerical-input-data/”. Are both for categorical target data feature selection using numerical data as they seem using the same data? If yes, why there are two posts with different methods for the same problem. Why there are two article with different methods? Am I missing something?!
I appreciate your time for your great tutorial which is my best resource.
I’m wondering how the score is calculated in f_classif method?
Hi Berkay…the following may be of interest:
https://datascience.stackexchange.com/questions/74465/how-to-understand-anova-f-for-feature-selection-in-python-sklearn-selectkbest-w
Hi Jason, Great website you’ve got there
How can I know whether a feature contributes towards each DV in a supervised ML model for example. Let’s say I predict the weight of some people in my data and I want to see if the “French Fries” feature aids to the model probability for someone with higher weight than for example the “Tomato” feature.
I have a bunch of features and want to know for each one if they contribute to the “0” or to the “1”
Thank you for your help
Hi Tal…The following may be of interest to you:
https://machinelearningmastery.com/an-introduction-to-feature-selection/
Hi, Is it a right way to use f_classif method to score the features with binary codes (0, 1), or (0, 1, 2, 3)?
For example a feature which shows people genders is coded by 0 and 1.
Thank you , very useful topic , I understand the concept very well
But I have a question , is it possible to apply PCA and Feature importance by DT , then take the common attributes between them . And use these attributes to apply ML model to increase the accuracy? Is there any methods to print the feature after applying PCA to dataset ?
Hi Jason Brownlee, Great website , and very informative !!
I have a question regarding how to choose the write feature selection algorithm for may problem ? I read one of your article in this regard. However, it only discuss how to choose filter methods, what about choosing the wrapper , embedded methods . How can I choose them , based on what ?
Than you very much Dr. James
Hi Carl…Thank you for your feedback and support! The following resource may be of interest to you:
https://machinelearningmastery.com/feature-selection-with-real-and-categorical-data/