This is a guest post by Igor Shvartser, a clever young student I have been coaching.
This post is part 1 in a 3 part series on modeling the famous Pima Indians Diabetes dataset that will introduce the problem and the data. Part 2 will investigate feature selection and spot checking algorithms and Part 3 in the series will investigate improvements to the classification accuracy and final presentation of results.
Kick-start your project with my new book Machine Learning Mastery With Weka, including step-by-step tutorials and clear screenshots for all examples.
Predict the Onset of Diabetes
Data mining and machine learning is helping medical professionals make diagnosis easier by bridging the gap between huge data sets and human knowledge. We can begin to apply machine learning techniques for classification in a dataset that describes a population that is under a high risk of the onset of diabetes.
Diabetes Mellitus affects 382 million people in the world, and the number of people with type-2 diabetes is increasing in every country. Untreated, diabetes can cause many complications.
Diabetes Test
Photo by Victor, some rights reserved.
The population for this study was the Pima Indian population near Phoenix, Arizona. The population has been under continuous study since 1965 by the National Institute of Diabetes and Digestive and Kidney Diseases because of its high incidence rate of diabetes.
For the purposes of this dataset, diabetes was diagnosed according to World Health Organization Criteria, which stated that if the 2 hour post-load glucose was at least 200 mg/dl at any survey exam or if the Indian Health Service Hospital serving the community found a glucose concentration of at least 200 mg/dl during the course of routine medical care.
Given the medical data we can gather about people, we should be able to make better predictions on how likely a person is to suffer the onset of diabetes, and therefore act appropriately to help. We can start analyzing data and experimenting with algorithms that will help us study the onset of diabetes in Pima Indians.
Need more help with Weka for Machine Learning?
Take my free 14-day email course and discover how to use the platform step-by-step.
Click to sign-up and also get a free PDF Ebook version of the course.
Related Work
Our study begins with an in-depth look of how researchers that used the same dataset approached the same problem. This helped me gain an understanding of the data and pave the way for my study, especially since the authors suggested alternative methods that are worth researching.
In 1988, Smith, Everhart, Dickson, Knowler, and Johannes performed an evaluation of using an early neural network model, ADAP, to forecast the onset of diabetes mellitus in a high-risk population of Pima Indians. They argued that the neural network approach would provide strong results when “the sample size is small, the form of underlying functional relationship is not known, and the underlying functional relationships involve complex interactions and intercorrelations among a number of variables“, see Using the ADAP Learning Algorithm to Forecast the Onset of Diabetes Mellitus.
They describe ADAP as “an adaptive learning routine that generates and executes digital analogs of perceptron-like devices“, see Learning in Control and Pattern Recognition Systems. The algorithm will make predictions based on a function of input variables, and will make internal adjustments if predictions are incorrect. The network is split into 3 main layers:
- Input, partitioned into “sensors”: Represents a discrete value. These are organized into partitions and are “excited” by input.
- Association Units: Uses a threshold function to activate a specific responder value. Connected to adjustable weights that change based on said function.
- Responder: Responder values are summed and constitute a specific prediction.
The network defined a “fixed matrix” that contained a partition for each attribute, a range of possible values, and the ability to identify connections in the data through a “variable array.” Rows in the matrix correspond to sensors, while columns correspond to association units. The variable array provided a way to easily identify connections between sensors and association units.
768 Females of Pima Indian Heritage
We can learn from the data found on UCI Machine Learning Repository which contains data on female patients at least 21 years old of Pima Indian heritage (update: download from here).
We have 768 instances and the following 8 attributes:
- Number of times pregnant (preg)
- Plasma glucose concentration a 2 hours in an oral glucose tolerance test (plas)
- Diastolic blood pressure in mm Hg (pres)
- Triceps skin fold thickness in mm (skin)
- 2-Hour serum insulin in mu U/ml (insu)
- Body mass index measured as weight in kg/(height in m)^2 (mass)
- Diabetes pedigree function (pedi)
- Age in years (age)
A particularly interesting attribute used in the study was the Diabetes Pedigree Function, pedi. It provided some data on diabetes mellitus history in relatives and the genetic relationship of those relatives to the patient. This measure of genetic influence gave us an idea of the hereditary risk one might have with the onset of diabetes mellitus. Based on observations in the proceeding section, it is unclear how well this function predicts the onset of diabetes.
Observations from Data
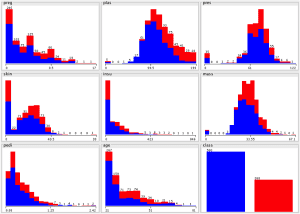
At first, I examined each attribute and reviewed the distribution parameters that Weka Explorer had prepared. I observed that:
- The preg and age attributes are integers.
- The population is generally young, less than 50 years old.
- Some attributes where a zero value exist seem to be errors in the data (e.g. plas, pres, skin, insu, and mass).
Upon examining the distribution of class values, I noticed that there are 500 negative instances (65.1%) and 258 positive instances (34.9%).
Reviewing histograms of all attributes in the dataset shows us that:
- Some of the attributes look normally distributed (plas, pres, skin, and mass).
- Some of the attributes look like they may have an exponential distribution (preg, insu, pedi, age).
- Age should probably have a normal distribution, the constraints on the data collection may have skewed the distribution.
- Testing for normality (normality plot) may be of interest. We could look at fitting the data to a normal distribution.
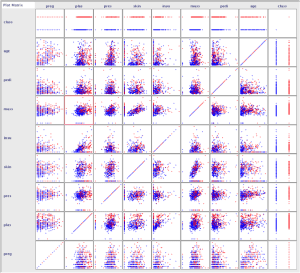
Reviewing scatter plots of all attributes in the dataset shows that:
- There is no obvious relationship between age and onset of diabetes.
- There is no obvious relationship between pedi function and onset of diabetes.
- This may suggest that diabetes is not hereditary, or that the Diabetes Pedigree Function needs work.
- Larger values of plas combined with larger values for age, pedi, mass, insu, skin, pres, and preg tends to show greater likelihood of testing positive for diabetes.
It is important to consider all possible limitations of the data, which may include the following:
- Results may be limited to Pima Indians, but give us a good start on how to begin diagnosing other populations with diabetes.
- Results may be limited to the time the data was collected (between 1960s and 1980s). Today’s medical procedures for diagnosing diabetes include a urine test and the hemoglobin A1c test, which shows the average level of blood sugar over the previous 3 months.
- Dataset is rather small, which may limit performance of some algorithms.
About Igor Shvartser
Hey! My name is Igor Shvartser and I’m studying math and computer science at the University of California in Santa Cruz. I’ve been really interested in machine learning, and was recently inspired to learn more after finishing a course called Machine Learning and Data Mining at my school.
I discovered the Machine Learning Mastery website, and found it really helpful, particularly with complementing the material in my course. Shortly after, I was excited to see that Jason reached out to me via email, and gave me an opportunity to contribute to his website. We were able to design a machine learning project that I’ll be working on over the next few weeks using Weka, a suite of machine learning software written at the University of Waikato. I hope to learn more about machine learning algorithms, applications, and data analysis, and will receive coaching from Jason whenever I may get stuck.
When I’m not learning about machine learning, I’m either mountain biking or working with a team to develop open source software that will let scientists analyze and collect data on images of molecular polymers. Before that, I was able to help change the way companies manage and understand their sustainability data within a small start-up in Santa Cruz.
Part 2
Stay tuned for Part 2 on feature selection and algorithm spot checking.






Another good article on your website, keep up good work!
Hi Igor and Jason,
Thanks for your efforts guys. 🙂 Here is my humble contribution.
– Reproduced the case study and explicitly stated the filters used
– Expanded on the case study by:
* balancing classes by over and under sampling minority and majority classes, respectively
* annotating predictions with probabilities
* exercising control over probability threshold to reduce false negatives at the cost of false positives
Github: https://github.com/dr-riz/diabetes
Well done!
After reproducing and expanding the case study in Weka, I decided to reproduce them in Python. Reasons:
I have developed understanding of the problem
Weka case will serve as a baseline for comparison
I want to get an idea on the amount of effort and flexibility both platforms provide on the same problem
For the warm up, I worked through Dr. Brownlee’s “Your First Machine Learning Project in Python Step-By-Step”. I wrote up the code with headings to allow the follower of the output to see what is going on.
In addition to my expanded Weka case study, the Python part expands the case study further by:
– additional algorithms for spot checking
– balancing class labels using LR parameter
– grid search on the hyperparameters
– searching for the crossover point between sensitivity and specificity
– plotting roc curve
– sensitivity and specificity cross over
– (not too exciting) saving and loading the model from disk
Github: https://github.com/dr-riz/diabetes
I like the way you explore the Pima India Dataset with many visualization and numerical checking !. Thank you.
In addition to the way you setup different models in order to compare their performance very elegantly (very useful for GridSearch, Spot check, benchmarking , etc.) …
I think one more time that Dr. Bronwlee tutorials inspire us a lot of, not only for main machine learning concepts but also for all the auxiliary tools needed such as Python, Keras, Sklearn, Matplot … many thanks to Jason !
JG
Thanks!
JG. I am glad you find my work on Pima dataset useful. 🙂
Well done!
Nice initiative. Kudos to both of you guys!
Hi, any clue how the diabetes pedigree was calculated?
Found it in https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2245318/pdf/procascamc00018-0276.pdf
Nice!
This might help:
https://github.com/jbrownlee/Datasets/blob/master/pima-indians-diabetes.names